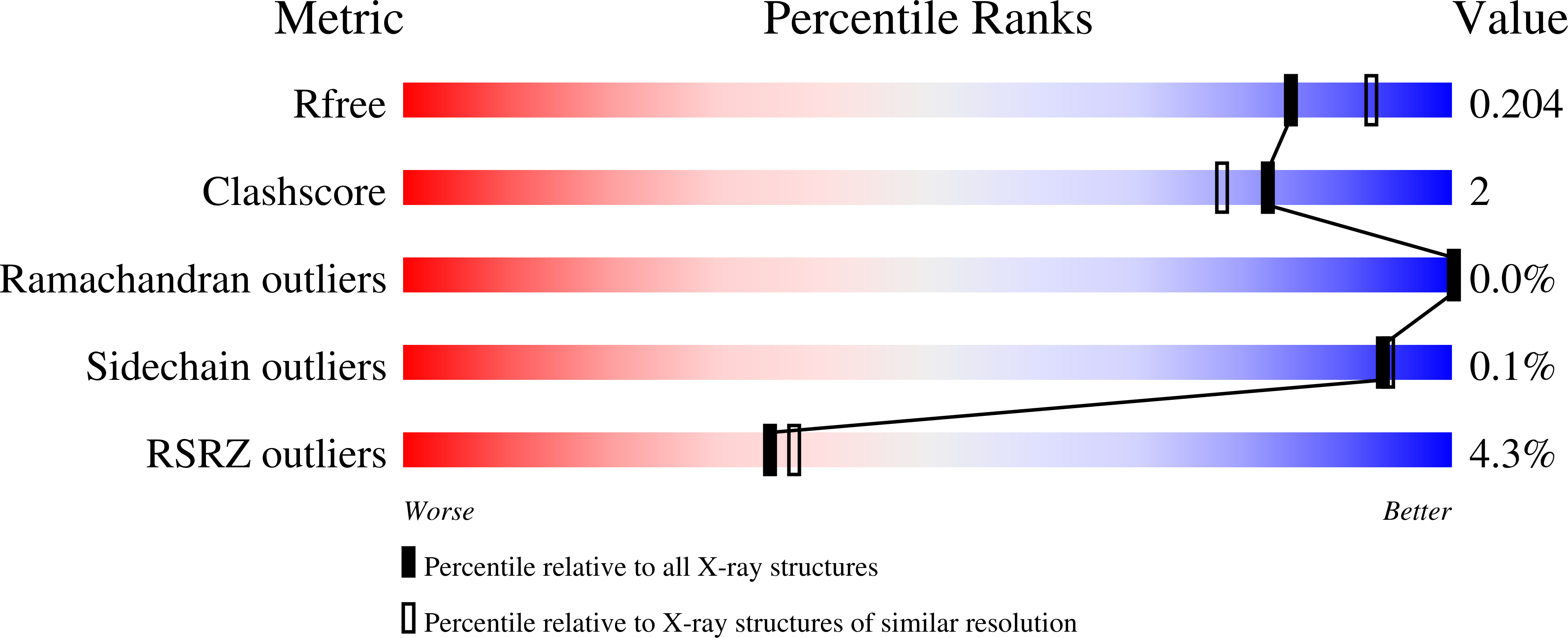
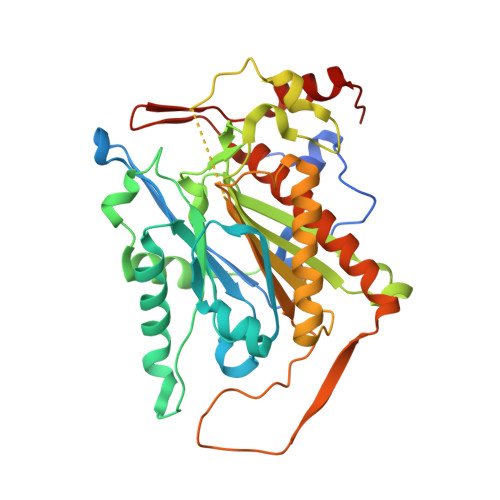
Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
John-White, M., Dumsday, G.J., Johanesen, P., Lyras, D., Drinkwater, N., McGowan, S.(2017) Acta Crystallogr F Struct Biol Commun 73: 386-392
- PubMed: 28695846
- DOI: https://doi.org/10.1107/S2053230X17007737
- Primary Citation of Related Structures:
5TZB - PubMed Abstract:
β-Aminopeptidases are a unique group of enzymes that have the unusual capability to hydrolyze N-terminal β-amino acids from synthetic β-peptides. β-Peptides can form secondary structures mimicking α-peptide-like structures that are resistant to degradation by most known proteases and peptidases. These characteristics of β-peptides give them great potential as peptidomimetics. Here, the X-ray crystal structure of BcA5-BapA, a β-aminopeptidase from a Gram-negative Burkholderia sp. that was isolated from activated sludge from a wastewater-treatment plant in Australia, is reported. The crystal structure of BcA5-BapA was determined to a resolution of 2.0 Å and showed a tetrameric assembly typical of the β-aminopeptidases. Each monomer consists of an α-subunit (residues 1-238) and a β-subunit (residues 239-367). Comparison of the structure of BcA5-BapA with those of other known β-aminopeptidases shows a highly conserved structure and suggests a similar proteolytic mechanism of action.
Organizational Affiliation:
Biomedicine Discovery Institute, Department of Microbiology, Monash University, Clayton, Melbourne, VIC 3800, Australia.