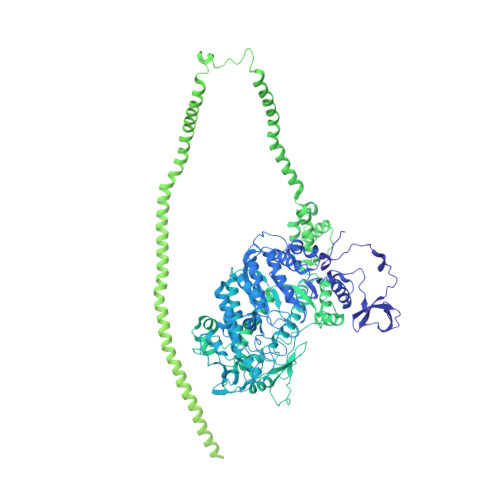
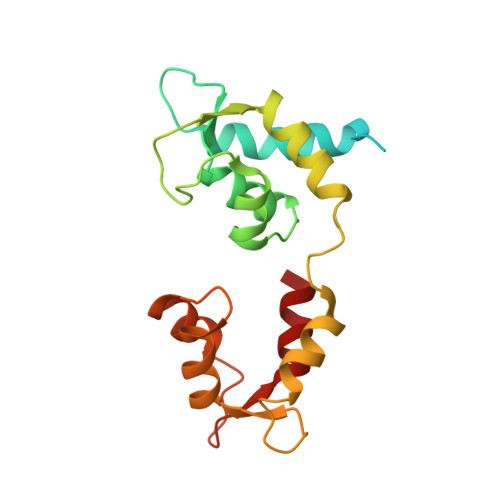
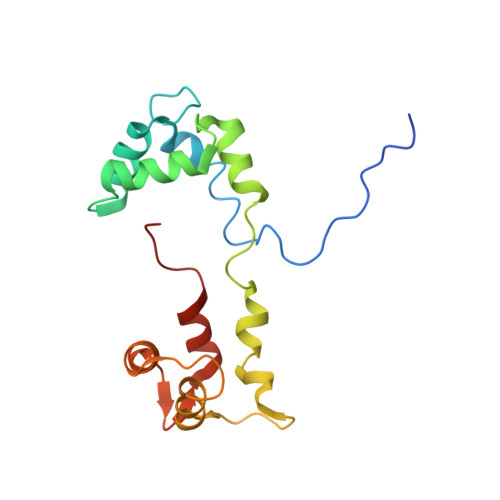
Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Alamo, L., Ware, J.S., Pinto, A., Gillilan, R.E., Seidman, J.G., Seidman, C.E., Padron, R.(2017) Elife 6
- PubMed: 28606303
- DOI: https://doi.org/10.7554/eLife.24634
- Primary Citation of Related Structures:
5TBY - PubMed Abstract:
Cardiac β-myosin variants cause hypertrophic (HCM) or dilated (DCM) cardiomyopathy by disrupting sarcomere contraction and relaxation. The locations of variants on isolated myosin head structures predict contractility effects but not the prominent relaxation and energetic deficits that characterize HCM. During relaxation, pairs of myosins form interacting-heads motif (IHM) structures that with other sarcomere proteins establish an energy-saving, super-relaxed (SRX) state. Using a human β-cardiac myosin IHM quasi-atomic model, we defined interactions sites between adjacent myosin heads and associated protein partners, and then analyzed rare variants from 6112 HCM and 1315 DCM patients and 33,370 ExAC controls. HCM variants, 72% that changed electrostatic charges, disproportionately altered IHM interaction residues (expected 23%; HCM 54%, p=2.6×10 -19 ; DCM 26%, p=0.66; controls 20%, p=0.23). HCM variant locations predict impaired IHM formation and stability, and attenuation of the SRX state - accounting for altered contractility, reduced diastolic relaxation, and increased energy consumption, that fully characterizes HCM pathogenesis.
Organizational Affiliation:
Centro de Biología Estructural, Instituto Venezolano de Investigaciones Científicas, Caracas, Venezuela.