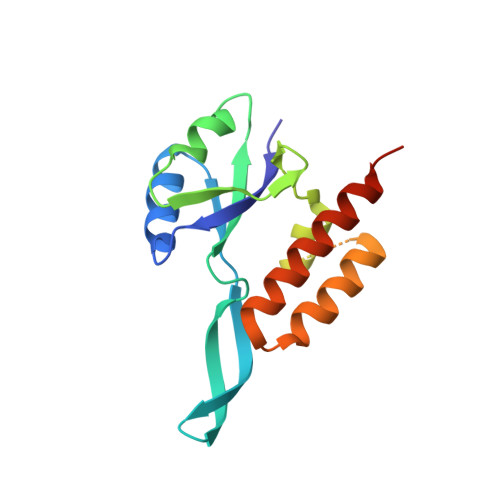
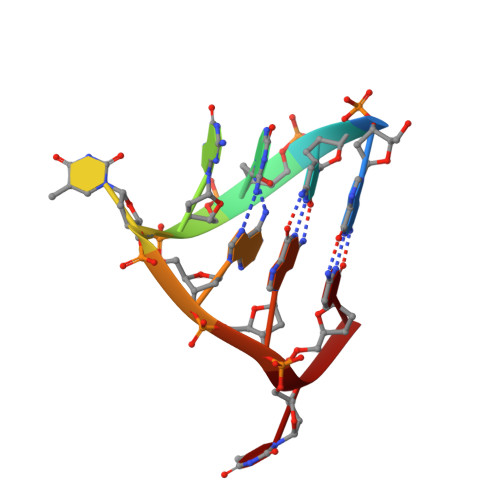
The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand.
Zuber, P.K., Artsimovitch, I., NandyMazumdar, M., Liu, Z., Nedialkov, Y., Schweimer, K., Rosch, P., Knauer, S.H.(2018) Elife 7
- PubMed: 29741479
- DOI: https://doi.org/10.7554/eLife.36349
- Primary Citation of Related Structures:
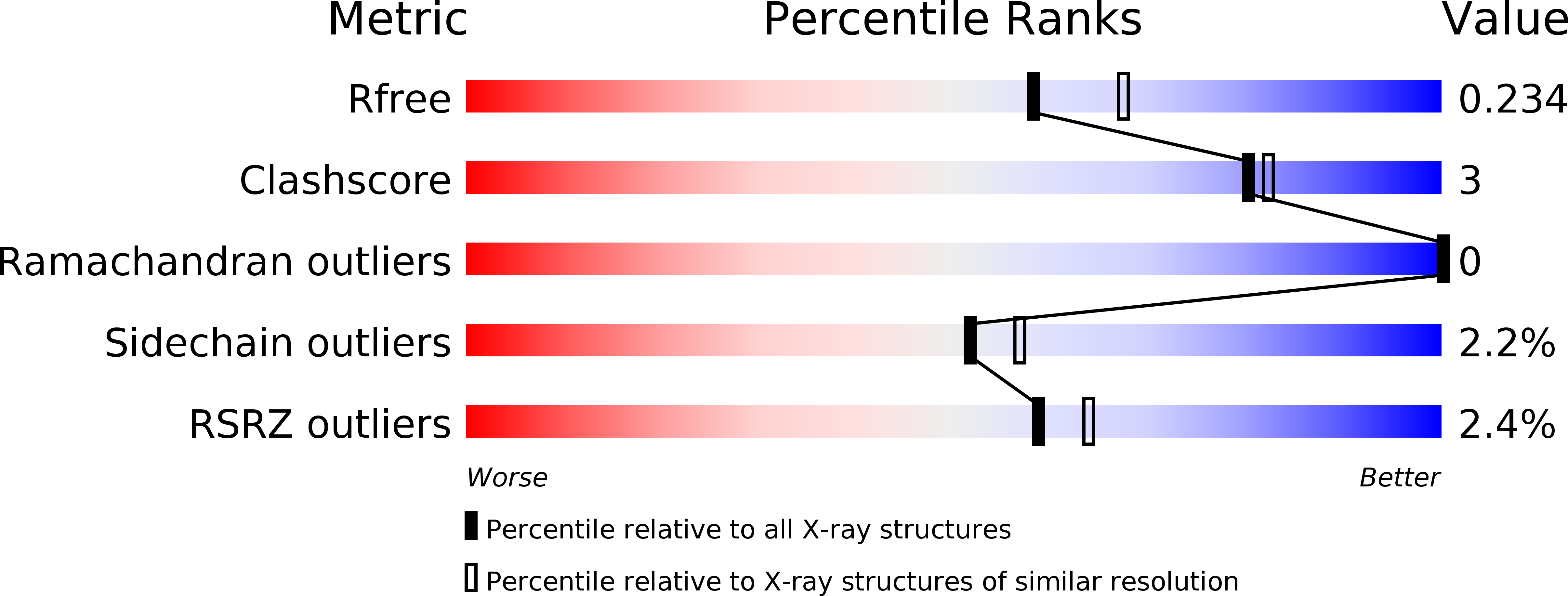
5OND - PubMed Abstract:
RfaH, a transcription regulator of the universally conserved NusG/Spt5 family, utilizes a unique mode of recruitment to elongating RNA polymerase to activate virulence genes. RfaH function depends critically on an ops sequence, an exemplar of a consensus pause, in the non-template DNA strand of the transcription bubble. We used structural and functional analyses to elucidate the role of ops in RfaH recruitment. Our results demonstrate that ops induces pausing to facilitate RfaH binding and establishes direct contacts with RfaH. Strikingly, the non-template DNA forms a hairpin in the RfaH: ops complex structure, flipping out a conserved T residue that is specifically recognized by RfaH. Molecular modeling and genetic evidence support the notion that ops hairpin is required for RfaH recruitment. We argue that both the sequence and the structure of the non-template strand are read out by transcription factors, expanding the repertoire of transcriptional regulators in all domains of life.
Organizational Affiliation:
Lehrstuhl Biopolymere und Forschungszentrum für Bio-Makromoleküle, Universität Bayreuth, Bayreuth, Germany.