Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Rengachari, S., Groiss, S., Devos, J.M., Caron, E., Grandvaux, N., Panne, D.(2018) Proc Natl Acad Sci U S A 115: E601-E609
- PubMed: 29317535
- DOI: https://doi.org/10.1073/pnas.1718426115
- Primary Citation of Related Structures:
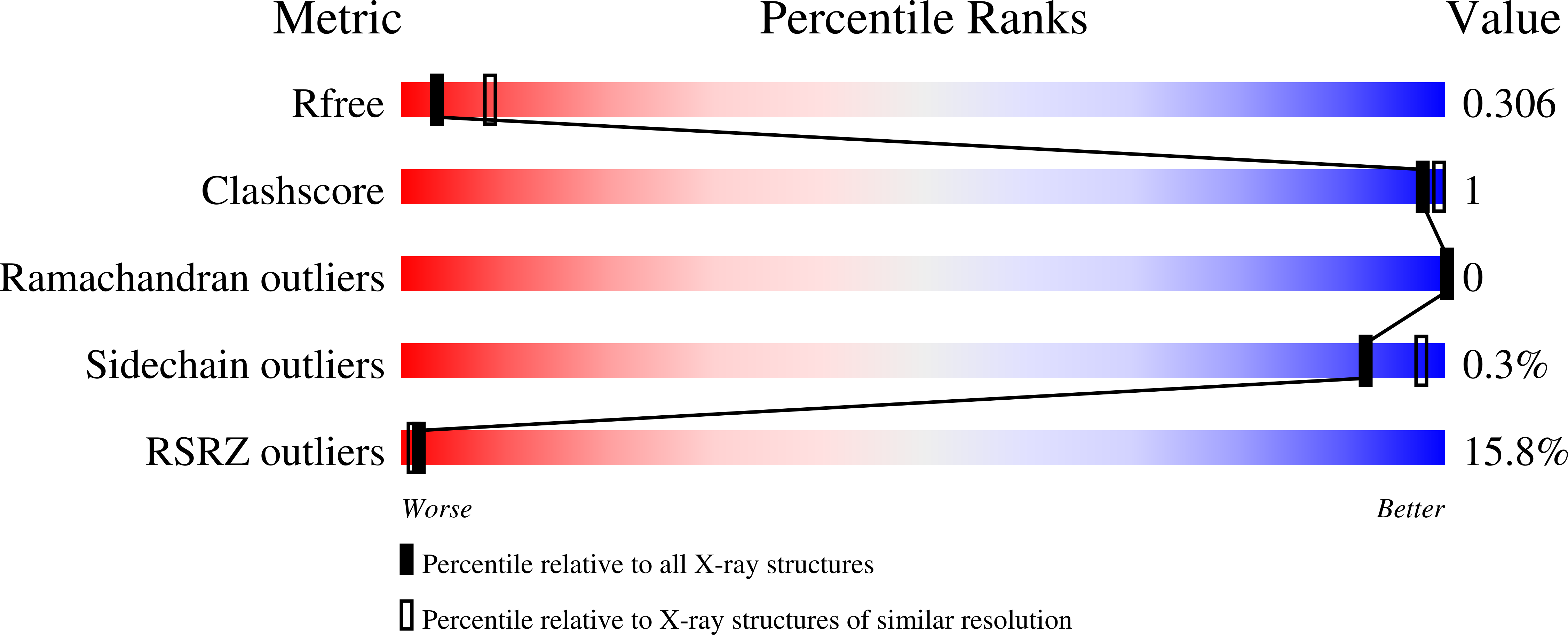
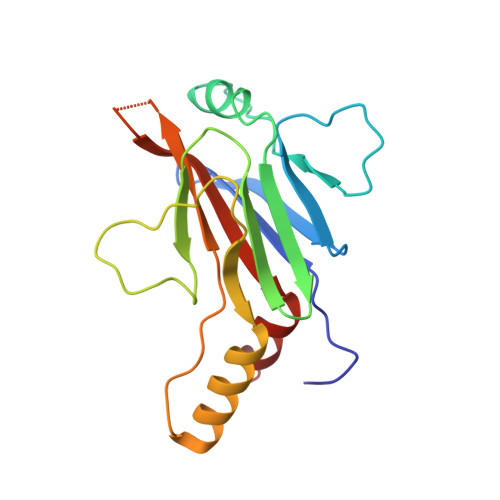
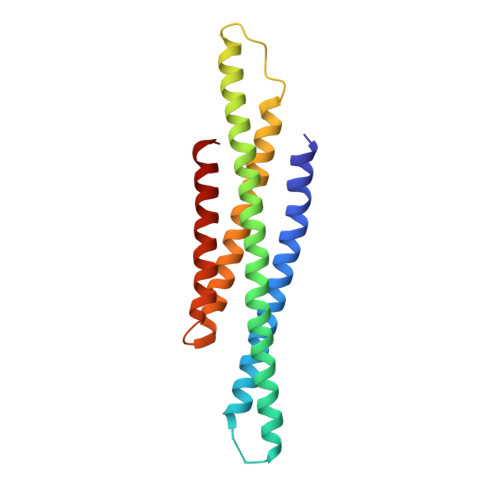
5OEM, 5OEN - PubMed Abstract:
Cytokine signaling through the JAK/STAT pathway controls multiple cellular responses including growth, survival, differentiation, and pathogen resistance. An expansion in the gene regulatory repertoire controlled by JAK/STAT signaling occurs through the interaction of STATs with IRF transcription factors to form ISGF3, a complex that contains STAT1, STAT2, and IRF9 and regulates expression of IFN-stimulated genes. ISGF3 function depends on selective interaction between IRF9, through its IRF-association domain (IAD), with the coiled-coil domain (CCD) of STAT2. Here, we report the crystal structures of the IRF9-IAD alone and in a complex with STAT2-CCD. Despite similarity in the overall structure among respective paralogs, the surface features of the IRF9-IAD and STAT2-CCD have diverged to enable specific interaction between these family members. We derive a model for the ISGF3 complex bound to an ISRE DNA element and demonstrate that the observed interface between STAT2 and IRF9 is required for ISGF3 function in cells.
Organizational Affiliation:
European Molecular Biology Laboratory, 38042 Grenoble, France.