Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Page, B.D.G., Valerie, N.C.K., Wright, R.H.G., Wallner, O., Isaksson, R., Carter, M., Rudd, S.G., Loseva, O., Jemth, A.S., Almlof, I., Font-Mateu, J., Llona-Minguez, S., Baranczewski, P., Jeppsson, F., Homan, E., Almqvist, H., Axelsson, H., Regmi, S., Gustavsson, A.L., Lundback, T., Scobie, M., Stromberg, K., Stenmark, P., Beato, M., Helleday, T.(2018) Nat Commun 9: 250-250
- PubMed: 29343827
- DOI: https://doi.org/10.1038/s41467-017-02293-7
- Primary Citation of Related Structures:
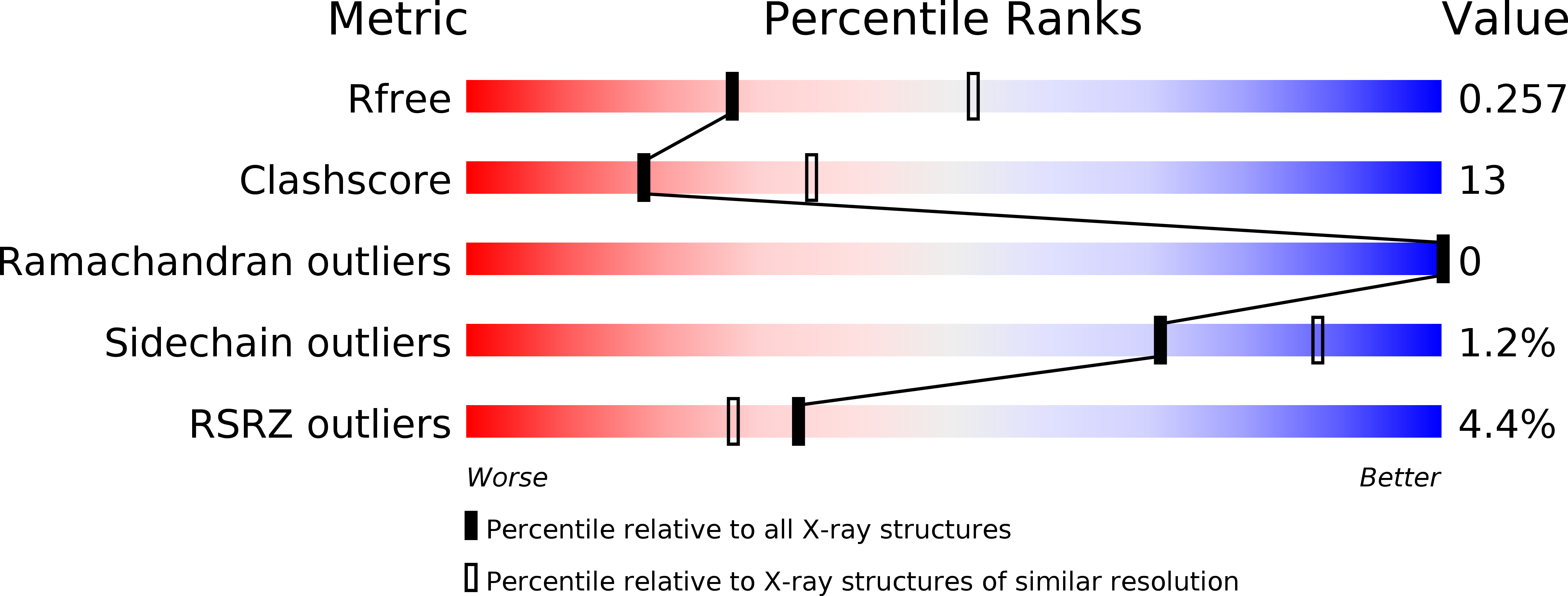
5NQR, 5NWH - PubMed Abstract:
With a diverse network of substrates, NUDIX hydrolases have emerged as a key family of nucleotide-metabolizing enzymes. NUDT5 (also called NUDIX5) has been implicated in ADP-ribose and 8-oxo-guanine metabolism and was recently identified as a rheostat of hormone-dependent gene regulation and proliferation in breast cancer cells. Here, we further elucidate the physiological relevance of known NUDT5 substrates and underscore the biological requirement for NUDT5 in gene regulation and proliferation of breast cancer cells. We confirm the involvement of NUDT5 in ADP-ribose metabolism and dissociate a relationship to oxidized nucleotide sanitation. Furthermore, we identify potent NUDT5 inhibitors, which are optimized to promote maximal NUDT5 cellular target engagement by CETSA. Lead compound, TH5427, blocks progestin-dependent, PAR-derived nuclear ATP synthesis and subsequent chromatin remodeling, gene regulation and proliferation in breast cancer cells. We herein present TH5427 as a promising, targeted inhibitor that can be used to further study NUDT5 activity and ADP-ribose metabolism.
Organizational Affiliation:
Science for Life Laboratory, Division of Translational Medicine and Chemical Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Solna, SE-171 21, Sweden. brent.page@scilifelab.se.