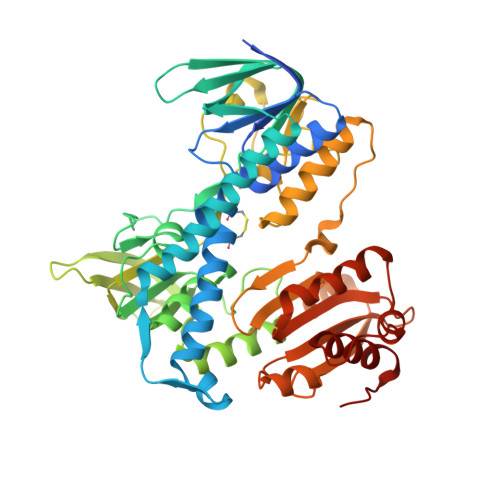
Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Szabo, E., Mizsei, R., Wilk, P., Zambo, Z., Torocsik, B., Weiss, M.S., Adam-Vizi, V., Ambrus, A.(2018) Free Radic Biol Med 124: 214-220
- PubMed: 29908278
- DOI: https://doi.org/10.1016/j.freeradbiomed.2018.06.008
- Primary Citation of Related Structures:
5J5Z, 5NHG - PubMed Abstract:
We report the crystal structures of the human (dihydro)lipoamide dehydrogenase (hLADH, hE3) and its disease-causing homodimer interface mutant D444V-hE3 at 2.27 and 1.84 Å resolution, respectively. The wild type structure is a unique uncomplexed, unliganded hE3 structure with the true canonical sequence. Based on the structural information a novel molecular pathomechanism is proposed for the impaired catalytic activity and enhanced capacity for reactive oxygen species generation of the pathogenic mutant. The mechanistic model involves a previously much ignored solvent accessible channel leading to the active site that might be perturbed also by other disease-causing homodimer interface substitutions of this enzyme.
Organizational Affiliation:
Department of Medical Biochemistry, MTA-SE Laboratory for Neurobiochemistry, Semmelweis University, H-1094 Budapest, Hungary.