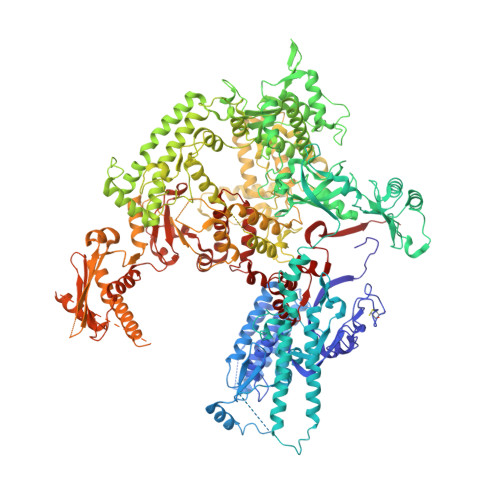
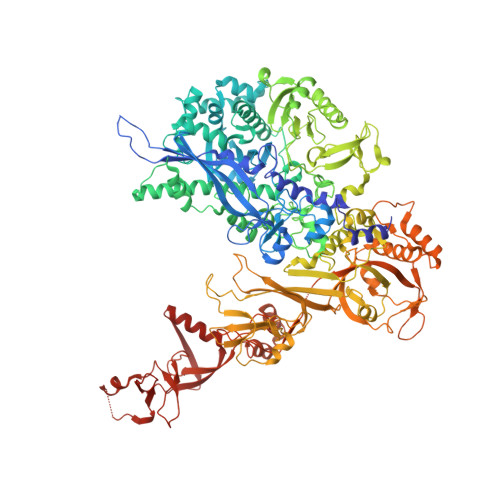
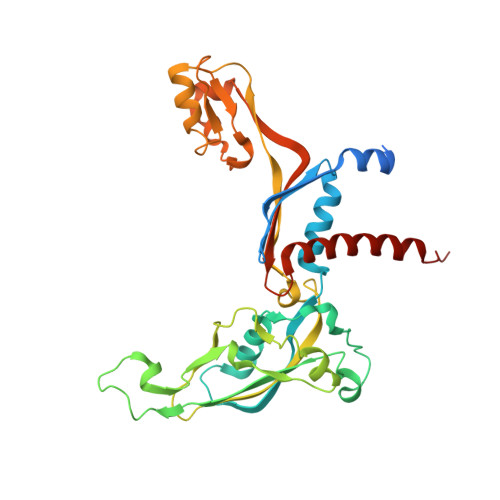
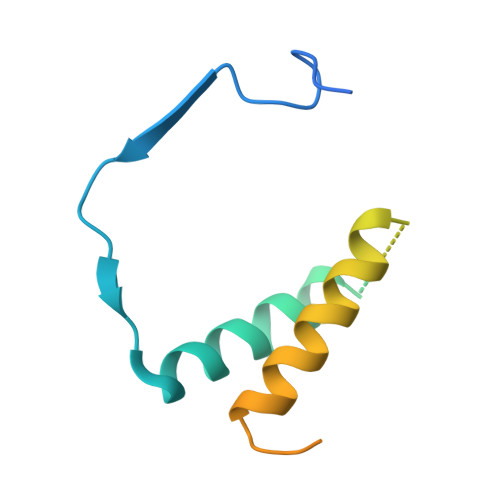
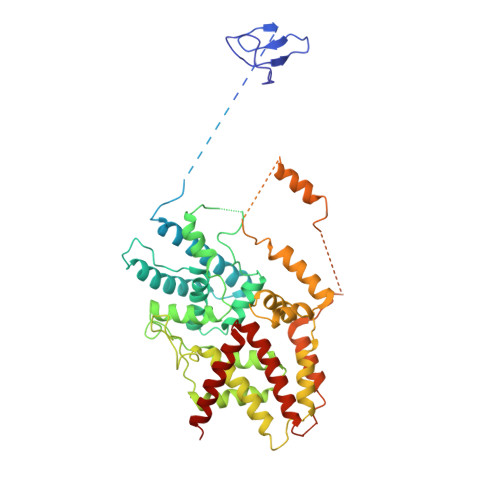
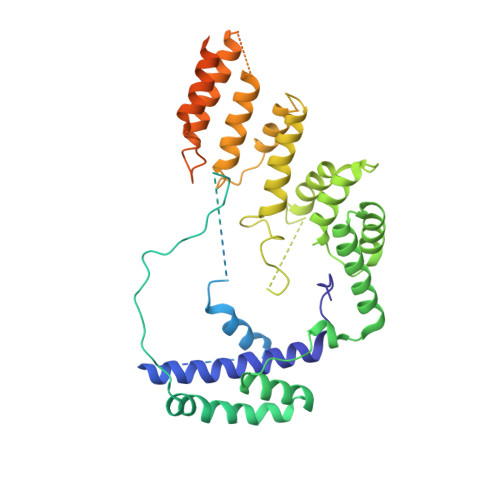
Structural Basis of RNA Polymerase I Transcription Initiation.
Engel, C., Gubbey, T., Neyer, S., Sainsbury, S., Oberthuer, C., Baejen, C., Bernecky, C., Cramer, P.(2017) Cell 169: 120-131.e22
- PubMed: 28340337
- DOI: https://doi.org/10.1016/j.cell.2017.03.003
- Primary Citation of Related Structures:
5N5Y, 5N5Z, 5N60, 5N61, 5O7X - PubMed Abstract:
Transcription initiation at the ribosomal RNA promoter requires RNA polymerase (Pol) I and the initiation factors Rrn3 and core factor (CF). Here, we combine X-ray crystallography and cryo-electron microscopy (cryo-EM) to obtain a molecular model for basal Pol I initiation. The three-subunit CF binds upstream promoter DNA, docks to the Pol I-Rrn3 complex, and loads DNA into the expanded active center cleft of the polymerase. DNA unwinding between the Pol I protrusion and clamp domains enables cleft contraction, resulting in an active Pol I conformation and RNA synthesis. Comparison with the Pol II system suggests that promoter specificity relies on a distinct "bendability" and "meltability" of the promoter sequence that enables contacts between initiation factors, DNA, and polymerase.
Organizational Affiliation:
Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.