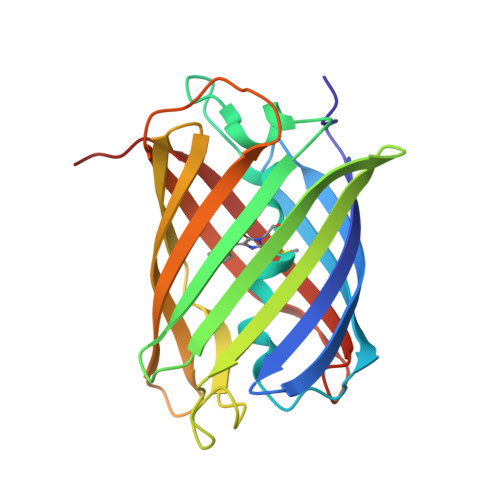
mScarlet: a bright monomeric red fluorescent protein for cellular imaging.
Bindels, D.S., Haarbosch, L., van Weeren, L., Postma, M., Wiese, K.E., Mastop, M., Aumonier, S., Gotthard, G., Royant, A., Hink, M.A., Gadella, T.W.(2017) Nat Methods 14: 53-56
- PubMed: 27869816
- DOI: https://doi.org/10.1038/nmeth.4074
- Primary Citation of Related Structures:
5LK4 - PubMed Abstract:
We report the engineering of mScarlet, a truly monomeric red fluorescent protein with record brightness, quantum yield (70%) and fluorescence lifetime (3.9 ns). We developed mScarlet starting with a consensus synthetic template and using improved spectroscopic screening techniques; mScarlet's crystal structure reveals a planar and rigidified chromophore. mScarlet outperforms existing red fluorescent proteins as a fusion tag, and it is especially useful as a Förster resonance energy transfer (FRET) acceptor in ratiometric imaging.
Organizational Affiliation:
Section of Molecular Cytology and van Leeuwenhoek Centre for Advanced Microscopy, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, the Netherlands.