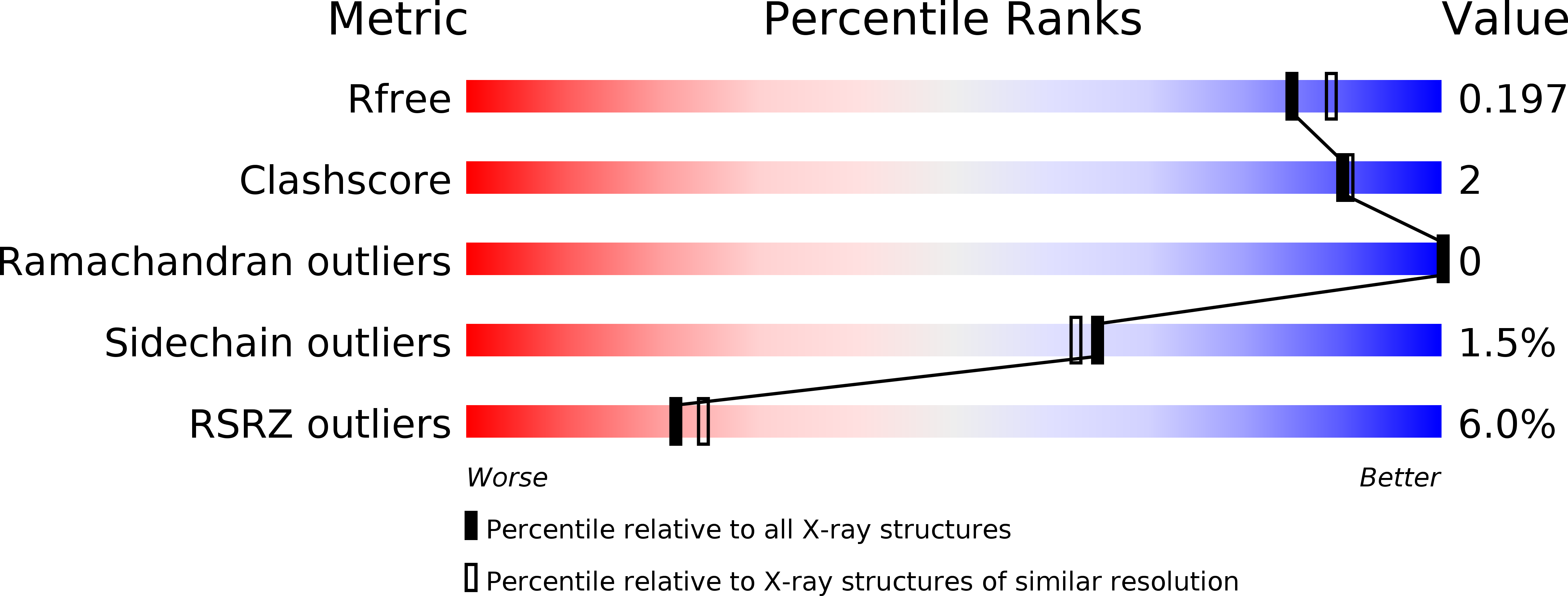
Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Pitsawong, W., Haynes, C.A., Koder, R.L., Rodgers, D.W., Miller, A.F.(2017) Structure 25: 978-987.e4
- PubMed: 28578873
- DOI: https://doi.org/10.1016/j.str.2017.05.002
- Primary Citation of Related Structures:
5J8D, 5J8G - PubMed Abstract:
Nitroreductase (NR) from Enterobacter cloacae reduces diverse nitroaromatics including herbicides, explosives, and prodrugs, and holds promise for bioremediation, prodrug activation, and enzyme-assisted synthesis. We solved crystal structures of NR complexes with bound substrate or analog for each of its two half-reactions. We complemented these with kinetic isotope effect (KIE) measurements elucidating H-transfer steps essential to each half-reaction. KIEs indicate hydride transfer from NADH to the flavin consistent with our structure of NR with the NADH analog nicotinic acid adenine dinucleotide (NAAD). The KIE on reduction of p-nitrobenzoic acid (p-NBA) also indicates hydride transfer, and requires revision of prior computational mechanisms. Our mechanistic information provided a structural restraint for the orientation of bound substrate, placing the nitro group closer to the flavin N5 in the pocket that binds the amide of NADH. KIEs show that solvent provides a proton, enabling accommodation of different nitro group placements, consistent with the broad repertoire of NR.
Organizational Affiliation:
Department of Chemistry, University of Kentucky, 505 Rose Street, Lexington, KY 40506-0055, USA.