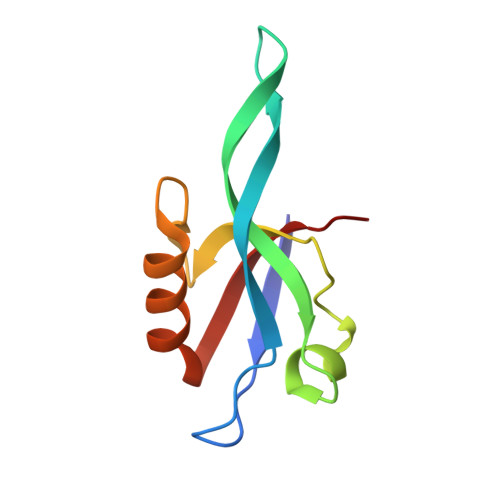
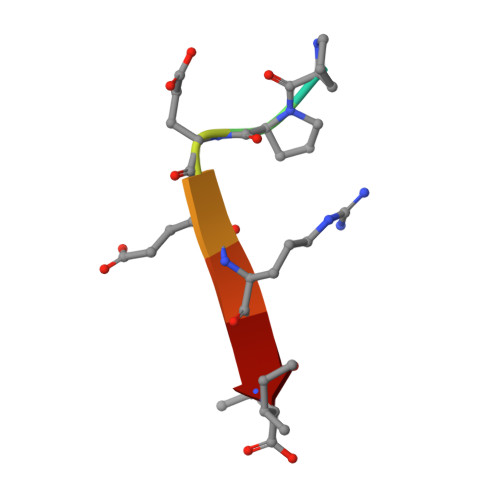
Binding of Crumbs to the Par-6 CRIB-PDZ Module Is Regulated by Cdc42.
Whitney, D.S., Peterson, F.C., Kittell, A.W., Egner, J.M., Prehoda, K.E., Volkman, B.F.(2016) Biochemistry 55: 1455-1461
- PubMed: 26894406
- DOI: https://doi.org/10.1021/acs.biochem.5b01342
- Primary Citation of Related Structures:
5I7Z - PubMed Abstract:
Par-6 is a scaffold protein that organizes other proteins into a complex required to initiate and maintain cell polarity. Cdc42-GTP binds the CRIB module of Par-6 and alters the binding affinity of the adjoining PDZ domain. Allosteric regulation of the Par-6 PDZ domain was first demonstrated using a peptide identified in a screen of typical carboxyl-terminal ligands. Crumbs, a membrane protein that localizes a conserved polarity complex, was subsequently identified as a functional partner for Par-6 that likely interacts with the PDZ domain. Here we show by nuclear magnetic resonance that Par-6 binds a Crumbs carboxyl-terminal peptide and report the crystal structure of the PDZ-peptide complex. The Crumbs peptide binds Par-6 more tightly than the previously studied carboxyl peptide ligand and interacts with the CRIB-PDZ module in a Cdc42-dependent manner. The Crumbs:Par-6 crystal structure reveals specific PDZ-peptide contacts that contribute to its higher affinity and Cdc42-enhanced binding. Comparisons with existing structures suggest that multiple C-terminal Par-6 ligands respond to a common conformational switch that transmits the allosteric effects of GTPase binding.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin , Milwaukee, Wisconsin 53226, United States.