Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Newman, J.A., Cooper, C.D., Roos, A.K., Aitkenhead, H., Oppermann, U.C., Cho, H.J., Osman, R., Gileadi, O.(2016) PLoS One 11: e0148762-e0148762
- PubMed: 26910052
- DOI: https://doi.org/10.1371/journal.pone.0148762
- Primary Citation of Related Structures:
2WA0, 4V0P, 5HVQ - PubMed Abstract:
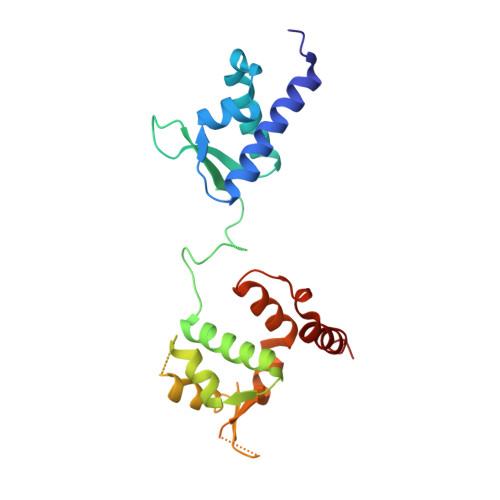
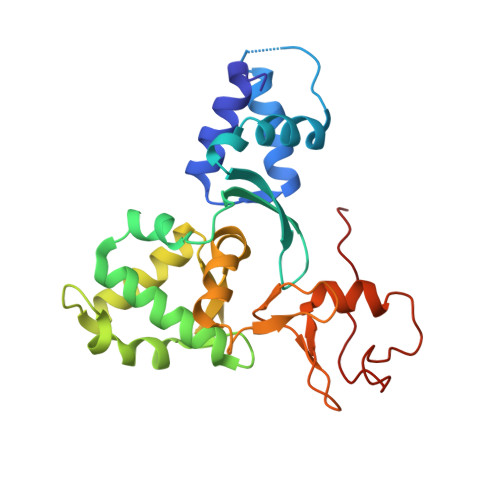
The MAGE (melanoma associated antigen) protein family are tumour-associated proteins normally present only in reproductive tissues such as germ cells of the testis. The human genome encodes over 60 MAGE genes of which one class (containing MAGE-A3 and MAGE-A4) are exclusively expressed in tumours, making them an attractive target for the development of targeted and immunotherapeutic cancer treatments. Some MAGE proteins are thought to play an active role in driving cancer, modulating the activity of E3 ubiquitin ligases on targets related to apoptosis. Here we determined the crystal structures of MAGE-A3 and MAGE-A4. Both proteins crystallized with a terminal peptide bound in a deep cleft between two tandem-arranged winged helix domains. MAGE-A3 (but not MAGE-A4), is predominantly dimeric in solution. Comparison of MAGE-A3 and MAGE-A3 with a structure of an effector-bound MAGE-G1 suggests that a major conformational rearrangement is required for binding, and that this conformational plasticity may be targeted by allosteric binders.
Organizational Affiliation:
Structural Genomics Consortium, University of Oxford, ORCRB, Roosevelt Drive, Oxford, OX3 7DQ, United Kingdom.