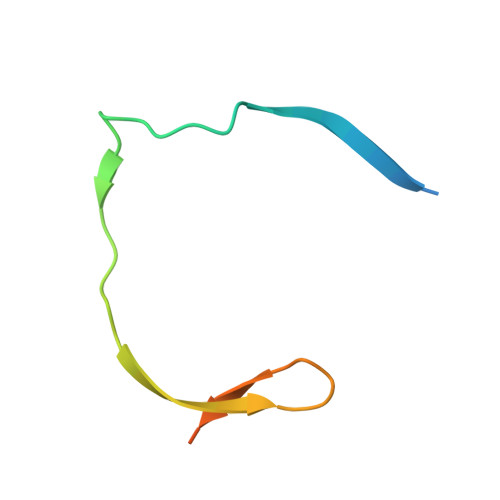
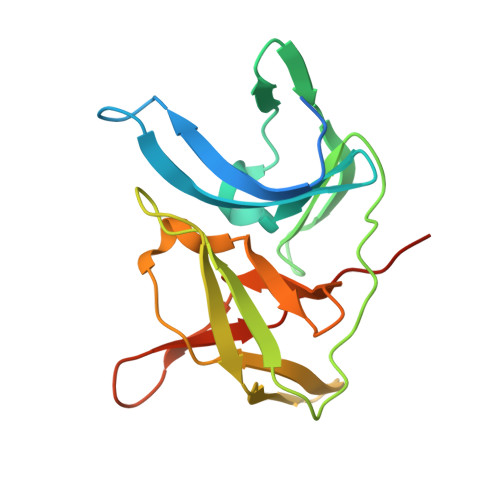
Crystal structure of unlinked NS2B-NS3 protease from Zika virus
Zhang, Z., Li, Y., Loh, Y.R., Phoo, W.W., Hung, A.W., Kang, C., Luo, D.(2016) Science 354: 1597-1600
- PubMed: 27940580
- DOI: https://doi.org/10.1126/science.aai9309
- Primary Citation of Related Structures:
5GPI, 5H4I - PubMed Abstract:
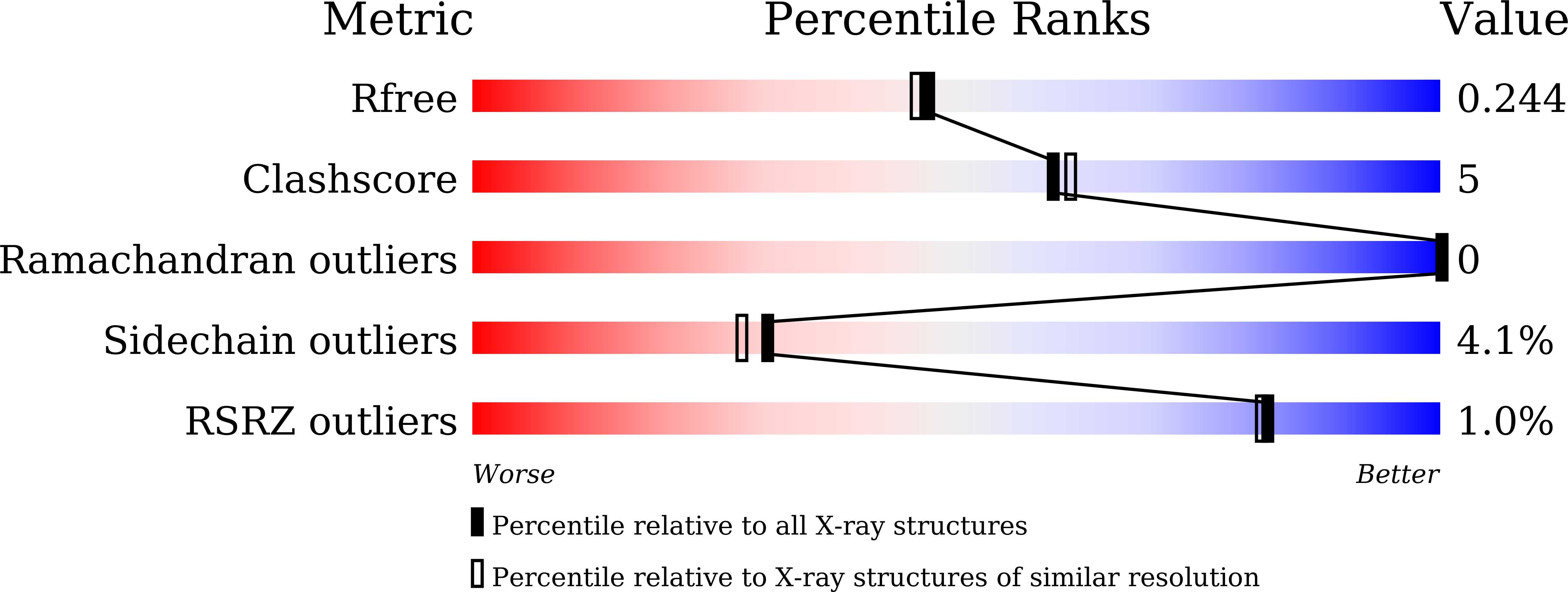
Zika virus (ZIKV) has rapidly emerged as a global public health concern. Viral NS2B-NS3 protease processes viral polyprotein and is essential for the virus replication, making it an attractive antiviral drug target. We report crystal structures at 1.58-angstrom resolution of the unlinked NS2B-NS3 protease from ZIKV as free enzyme and bound to a peptide reversely oriented at the active site. The unlinked NS2B-NS3 protease adopts a closed conformation in which NS2B engages NS3 to form an empty substrate-binding site. A second protease in the same crystal binds to the residues K14K15G16E17 from the neighboring NS3 in reverse orientation, resisting proteolysis. These features of ZIKV NS2B-NS3 protease may accelerate the discovery of structure-based antiviral drugs against ZIKV and related pathogenic flaviviruses.
Organizational Affiliation:
Lee Kong Chian School of Medicine, Nanyang Technological University, Experimental Medicine Building 03-07, 59 Nanyang Drive, Singapore 636921.