Structural basis for substrate binding and catalysis of branching enzyme from Cyanothece sp. ATCC 51142
Hayashi, M., Suzuki, R., Colleoni, C., Ball, S.G., Fujita, N., Suzuki, E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 1,4-alpha-glucan branching enzyme GlgB | 793 | Crocosphaera subtropica ATCC 51142 | Mutation(s): 1 Gene Names: glgB, glgB1, cce_2248 EC: 2.4.1.18 | 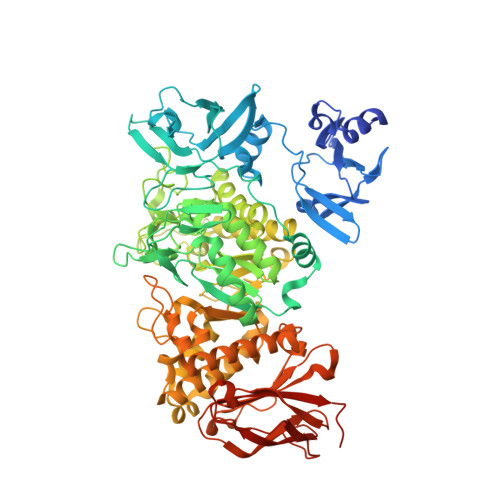 | |
UniProt | |||||
Find proteins for B1WPM8 (Crocosphaera subtropica (strain ATCC 51142 / BH68)) Explore B1WPM8 Go to UniProtKB: B1WPM8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B1WPM8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A], C [auth A], D [auth A], E [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MG Query on MG | F [auth A], G [auth A], H [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 133.87 | α = 90 |
| b = 133.87 | β = 90 |
| c = 185.01 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Shimadzu Science Foundation | Japan | 120003 |
| JSPS KAKENHI | Japan | 15K18685 |
| JSPS KAKENHI | Japan | 25440193 |