Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
Dousson, C., Alexandre, F.R., Amador, A., Bonaric, S., Bot, S., Caillet, C., Convard, T., da Costa, D., Lioure, M.P., Roland, A., Rosinovsky, E., Maldonado, S., Parsy, C., Trochet, C., Storer, R., Stewart, A., Wang, J., Mayes, B.A., Musiu, C., Poddesu, B., Vargiu, L., Liuzzi, M., Moussa, A., Jakubik, J., Hubbard, L., Seifer, M., Standring, D.(2016) J Med Chem 59: 1891-1898
- PubMed: 26804933
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01430
- Primary Citation of Related Structures:
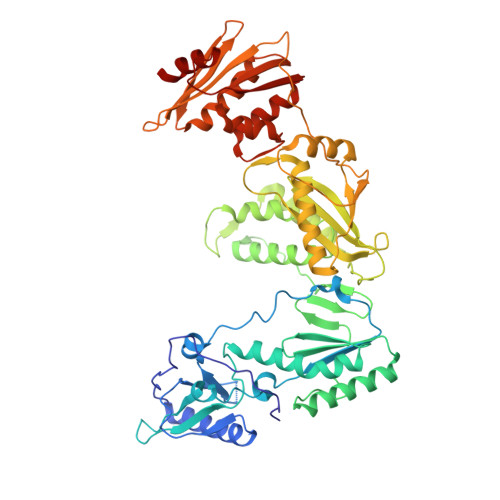
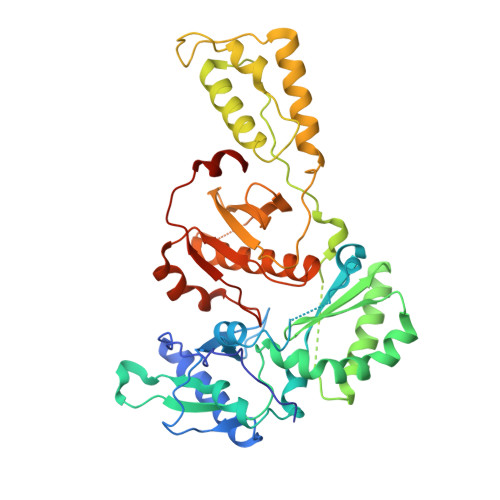
5FDL - PubMed Abstract:
Here, we describe the design, synthesis, biological evaluation, and identification of a clinical candidate non-nucleoside reverse transcriptase inhibitors (NNRTIs) with a novel aryl-phospho-indole (APhI) scaffold. NNRTIs are recommended components of highly active antiretroviral therapy (HAART) for the treatment of HIV-1. Since a major problem associated with NNRTI treatment is the emergence of drug resistant virus, this work focused on optimization of the APhI against clinically relevant HIV-1 Y181C and K103N mutants and the Y181C/K103N double mutant. Optimization of the phosphinate aryl substituent led to the discovery of the 3-Me,5-acrylonitrile-phenyl analogue RP-13s (IDX899) having an EC50 of 11 nM against the Y181C/K103N double mutant.
Organizational Affiliation:
IDENIX, an MSD Company , 1682 rue de la Valsière, Cap Gamma, BP 50001, 34189 Cedex 4 Montpellier, France.