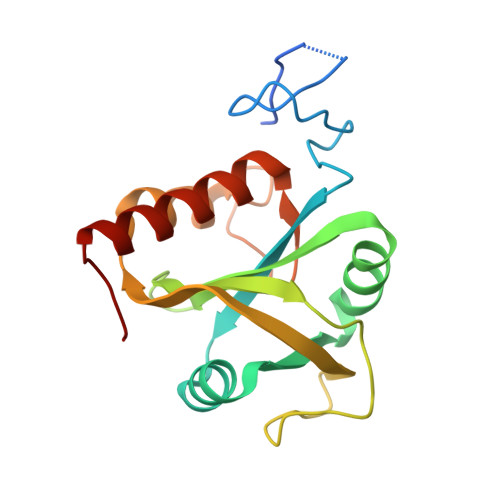
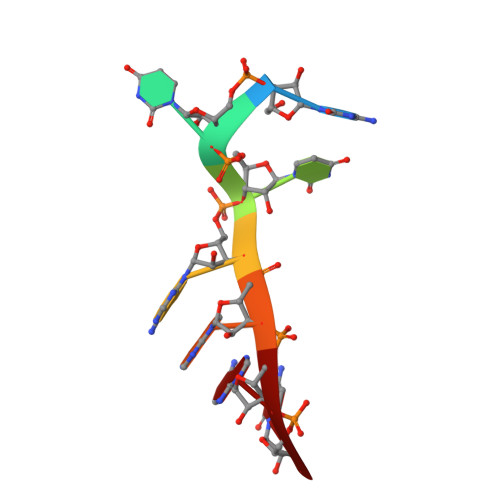
A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1
Wang, C.Y., Zhu, Y.W., Bao, H.Y., Jiang, Y.Y., Xu, C., Wu, J.H., Shi, Y.Y.(2016) Nucleic Acids Res 44: 969-982
- PubMed: 26673708
- DOI: https://doi.org/10.1093/nar/gkv1382
- Primary Citation of Related Structures:
5DNO, 5DNP - PubMed Abstract:
The YTH domain-containing protein Mmi1, together with other factors, constitutes the machinery used to selectively remove meiosis-specific mRNA during the vegetative growth of fission yeast. Mmi1 directs meiotic mRNAs to the nuclear exosome for degradation by recognizing their DSR (determinant of selective removal) motif. Here, we present the crystal structure of the Mmi1 YTH domain in the apo state and in complex with a DSR motif, demonstrating that the Mmi1 YTH domain selectively recognizes the DSR motif. Intriguingly, Mmi1 also contains a potential m(6)A (N(6)-methyladenine)-binding pocket, but its binding of the DSR motif is dependent on a long groove opposite the m(6)A pocket. The DSR-binding mode is distinct from the m(6)A RNA-binding mode utilized by other YTH domains. Furthermore, the m(6)A pocket cannot bind m(6)A RNA. Our structural and biochemical experiments uncover the mechanism of the YTH domain in binding the DSR motif and help to elucidate the function of Mmi1.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Anhui 230027, China.