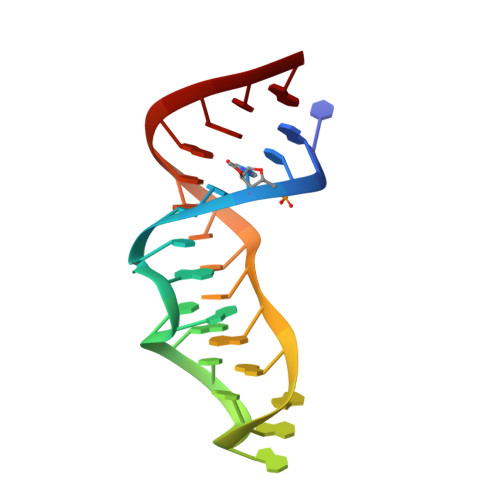
Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.
Mooers, B.H.(2016) Acta Crystallogr D Struct Biol 72: 477-487
- PubMed: 27050127
- DOI: https://doi.org/10.1107/S2059798316001224
- Primary Citation of Related Structures:
5D99, 5DA6 - PubMed Abstract:
Using direct methods starting from random phases, the crystal structure of a 32-base-pair RNA (675 non-H RNA atoms in the asymmetric unit) was determined using only the native diffraction data (resolution limit 1.05 Å) and the computer program SIR2014. The almost three helical turns of the RNA in the asymmetric unit introduced partial or imperfect translational pseudosymmetry (TPS) that modulated the intensities when averaged by the l Miller indices but still escaped automated detection. Almost six times as many random phase sets had to be tested on average to reach a correct structure compared with a similar-sized RNA hairpin (27 nucleotides, 580 non-H RNA atoms) without TPS. More sensitive methods are needed for the automated detection of partial TPS.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, and Stephenson Cancer Center, University of Oklahoma Health Sciences Center, 975 NE 10th Street, BRC 466, Oklahoma City, OK 73104, USA.