Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Bauman, J.D., Harrison, J.J.E.K., Arnold, E.(2016) IUCrJ 3: 51-60
Experimental Data Snapshot
(2016) IUCrJ 3: 51-60
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HIV-1 reverse transcriptase, p66 subunit | 557 | Human immunodeficiency virus type 1 BH10 | Mutation(s): 3 EC: 2.7.7.49 | 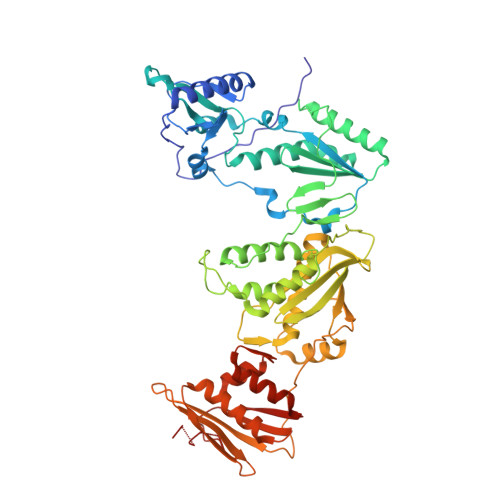 | |
UniProt | |||||
Find proteins for P03366 (Human immunodeficiency virus type 1 group M subtype B (isolate BH10)) Explore P03366 Go to UniProtKB: P03366 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03366 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HIV-1 reverse transcriptase, p51 subunit | 428 | Human immunodeficiency virus type 1 BH10 | Mutation(s): 1 EC: 2.7.7.49 | 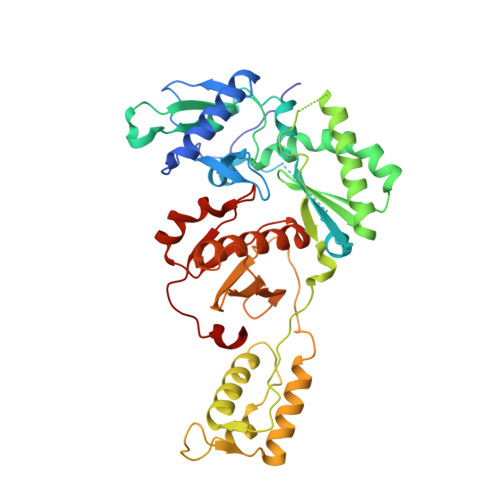 | |
UniProt | |||||
Find proteins for P03366 (Human immunodeficiency virus type 1 group M subtype B (isolate BH10)) Explore P03366 Go to UniProtKB: P03366 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03366 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| T27 Query on T27 | C [auth A] | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile C22 H18 N6 YIBOMRUWOWDFLG-ONEGZZNKSA-N |  | ||
| PYZ Query on PYZ | CA [auth B] DA [auth B] EA [auth B] FA [auth B] S [auth A] | 4-IODOPYRAZOLE C3 H3 I N2 LLNQWPTUJJYTTE-UHFFFAOYSA-N |  | ||
| IOD Query on IOD | AA [auth B] BA [auth B] I [auth A] J [auth A] K [auth A] | IODIDE ION I XMBWDFGMSWQBCA-UHFFFAOYSA-M |  | ||
| SO4 Query on SO4 | D [auth A], E [auth A], F [auth A], V [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| DMS Query on DMS | GA [auth B], HA [auth B], IA [auth B], JA [auth B], U [auth A] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | G [auth A], H [auth A], W [auth B], X [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 162.842 | α = 90 |
| b = 73.483 | β = 100.67 |
| c = 109.582 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PHENIX | phasing |