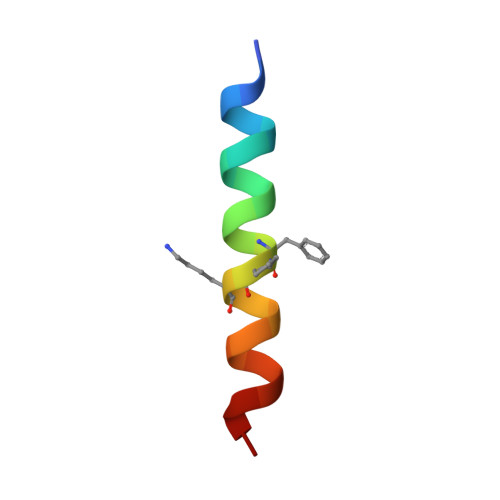
Quasiracemate Crystal Structures of Magainin 2 Derivatives Support the Functional Significance of the Phenylalanine Zipper Motif.
Hayouka, Z., Thomas, N.C., Mortenson, D.E., Satyshur, K.A., Weisblum, B., Forest, K.T., Gellman, S.H.(2015) J Am Chem Soc 137: 11884-11887
- PubMed: 26369301
- DOI: https://doi.org/10.1021/jacs.5b07206
- Primary Citation of Related Structures:
5CGN, 5CGO - PubMed Abstract:
Quasiracemic crystallography has been used to explore the significance of homochiral and heterochiral associations in a set of host-defense peptide derivatives. The previously reported racemic crystal structure of a magainin 2 derivative displayed a homochiral antiparallel dimer association featuring a "phenylalanine zipper" notable for the dual roles of phenylalanines in mediating dimerization and formation of an exposed hydrophobic swath. This motif is seen as well in two new quasiracemate crystals that contain the d form of the magainin 2 derivative along with an l-peptide in which one Ala has been replaced by a β-amino acid residue. This structural trend supports the hypothesis that the Phe zipper motif has functional significance.
Organizational Affiliation:
Department of Chemistry, ‡Department of Medicine, and §Department of Bacteriology, University of Wisconsin-Madison , Madison, Wisconsin 53706, United States.