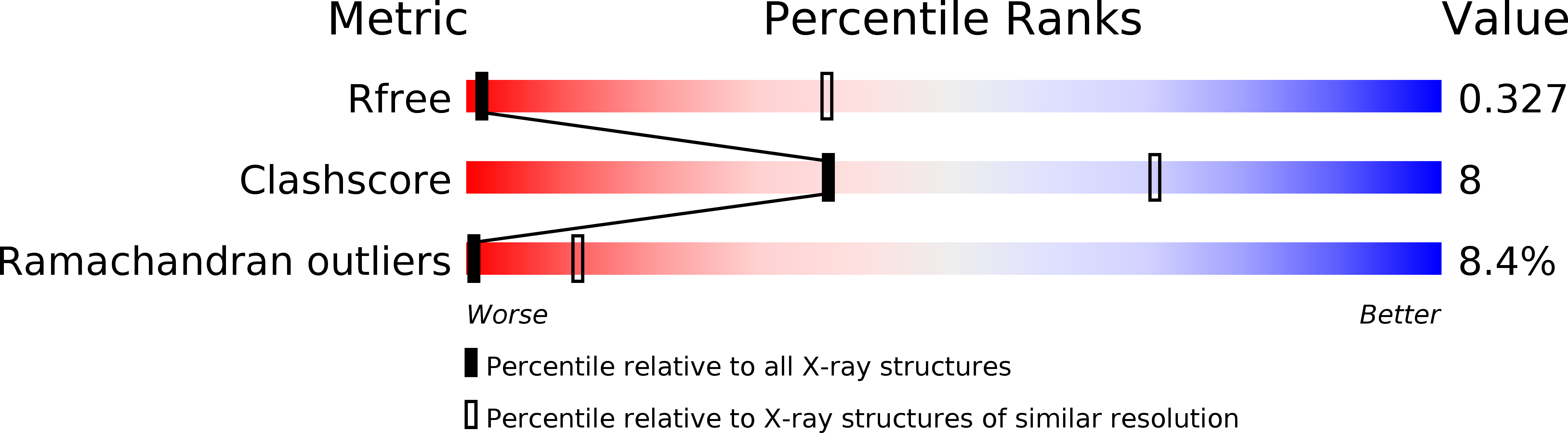Structure and mechanism of an active lipid-linked oligosaccharide flippase.
Perez, C., Gerber, S., Boilevin, J., Bucher, M., Darbre, T., Aebi, M., Reymond, J.L., Locher, K.P.(2015) Nature 524: 433-438
- PubMed: 26266984
- DOI: https://doi.org/10.1038/nature14953
- Primary Citation of Related Structures:
5C73, 5C76, 5C78 - PubMed Abstract:
The flipping of membrane-embedded lipids containing large, polar head groups is slow and energetically unfavourable, and is therefore catalysed by flippases, the mechanisms of which are unknown. A prominent example of a flipping reaction is the translocation of lipid-linked oligosaccharides that serve as donors in N-linked protein glycosylation. In Campylobacter jejuni, this process is catalysed by the ABC transporter PglK. Here we present a mechanism of PglK-catalysed lipid-linked oligosaccharide flipping based on crystal structures in distinct states, a newly devised in vitro flipping assay, and in vivo studies. PglK can adopt inward- and outward-facing conformations in vitro, but only outward-facing states are required for flipping. While the pyrophosphate-oligosaccharide head group of lipid-linked oligosaccharides enters the translocation cavity and interacts with positively charged side chains, the lipidic polyprenyl tail binds and activates the transporter but remains exposed to the lipid bilayer during the reaction. The proposed mechanism is distinct from the classical alternating-access model applied to other transporters.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.