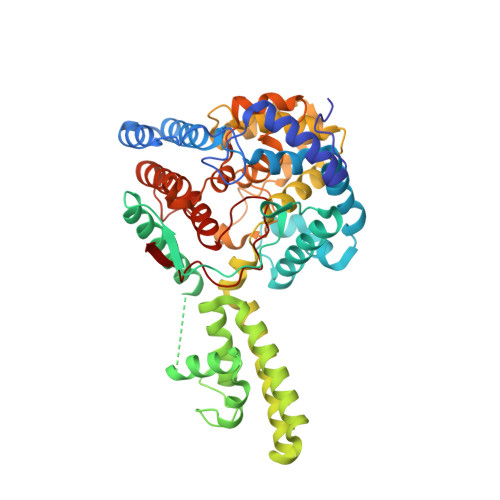
The Crystal Structure of the Hazara Virus Nucleocapsid Protein.
Surtees, R., Ariza, A., Punch, E.K., Trinh, C.H., Dowall, S.D., Hewson, R., Hiscox, J.A., Barr, J.N., Edwards, T.A.(2015) BMC Struct Biol 15: 24
- PubMed: 26715309
- DOI: https://doi.org/10.1186/s12900-015-0051-3
- Primary Citation of Related Structures:
5A97 - PubMed Abstract:
Hazara virus (HAZV) is a member of the Bunyaviridae family of segmented negative stranded RNA viruses, and shares the same serogroup as Crimean-Congo haemorrhagic fever virus (CCHFV). CCHFV is responsible for fatal human disease with a mortality rate approaching 30 %, which has an increased recent incidence within southern Europe. There are no preventative or therapeutic treatments for CCHFV-mediated disease, and thus CCHFV is classified as a hazard group 4 pathogen. In contrast HAZV is not associated with serious human disease, although infection of interferon receptor knockout mice with either CCHFV or HAZV results in similar disease progression. To characterise further similarities between HAZV and CCHFV, and support the use of HAZV as a model for CCHFV infection, we investigated the structure of the HAZV nucleocapsid protein (N) and compared it to CCHFV N. N performs an essential role in the viral life cycle by encapsidating the viral RNA genome, and thus, N represents a potential therapeutic target.
Organizational Affiliation:
Public Health England, Porton Down, Salisbury, Wiltshire, SP4 0JG, UK. bs06ras@leeds.ac.uk.