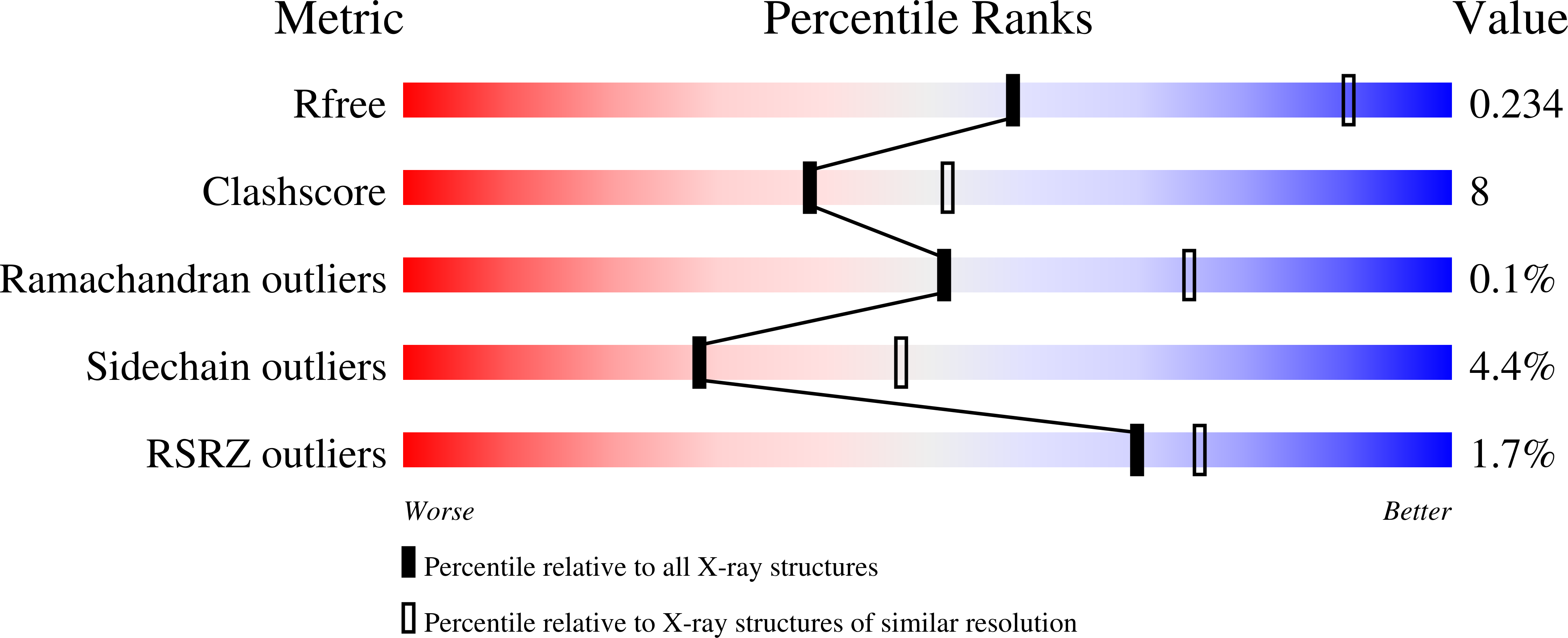
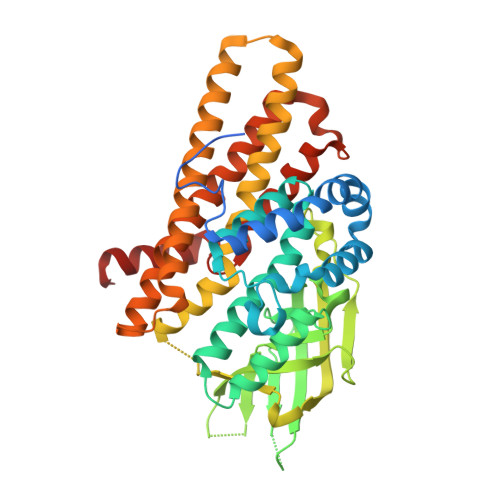
Structure of DnmZ, a nitrososynthase in the Streptomyces peucetius anthracycline biosynthetic pathway.
Sartor, L., Ibarra, C., Al-Mestarihi, A., Bachmann, B.O., Vey, J.L.(2015) Acta Crystallogr F Struct Biol Commun 71: 1205-1214
- PubMed: 26457508
- DOI: https://doi.org/10.1107/S2053230X15014272
- Primary Citation of Related Structures:
4ZXV, 4ZYJ - PubMed Abstract:
The anthracyclines are a class of highly effective natural product chemotherapeutics and are used to treat a range of cancers, including leukemia. The toxicity of the anthracyclines has stimulated efforts to further diversify the scaffold of the natural product, which has led to renewed interest in the biosynthetic pathway responsible for the formation and modification of this family of molecules. DnmZ is an N-hydroxylating flavin monooxygenase (a nitrososynthase) that catalyzes the oxidation of the exocyclic amine of the sugar nucleotide dTDP-L-epi-vancosamine to its nitroso form. Its specific role in the anthracycline biosynthetic pathway involves the synthesis of the seven-carbon acetal moiety attached to C4 of L-daunosamine observed in the anthracycline baumycin. Here, X-ray crystallography was used to elucidate the three-dimensional structure of DnmZ. Two crystal structures of DnmZ were yielded: that of the enzyme alone, solved to 3.00 Å resolution, and that of the enzyme in complex with thymidine diphosphate, the nucleotide carrier portion of the substrate, solved to 2.74 Å resolution. These models add insights into the structural features involved in substrate specificity and conformational changes involved in thymidine diphosphate binding by the nitrososynthases.
Organizational Affiliation:
Department of Chemistry and Biochemistry, California State University Northridge, Northridge, CA 91330, USA.