Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763)
Arbing, M.A., Chan, S., Kuo, E., Harris, L.R., Zhou, T.T., Eisenberg, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 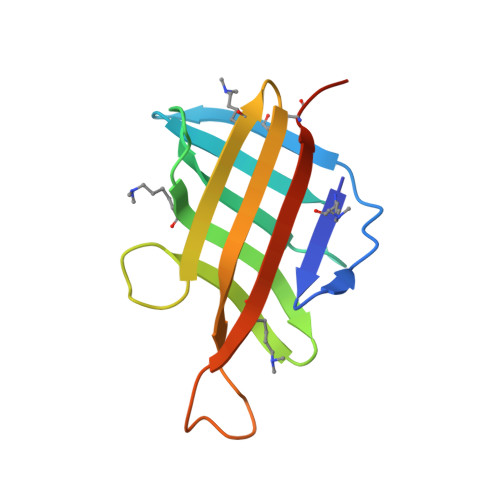 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 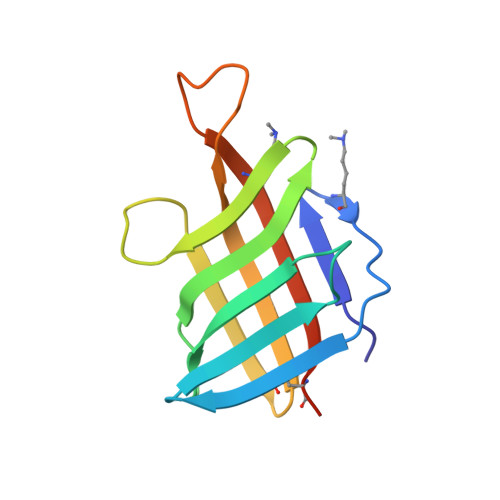 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 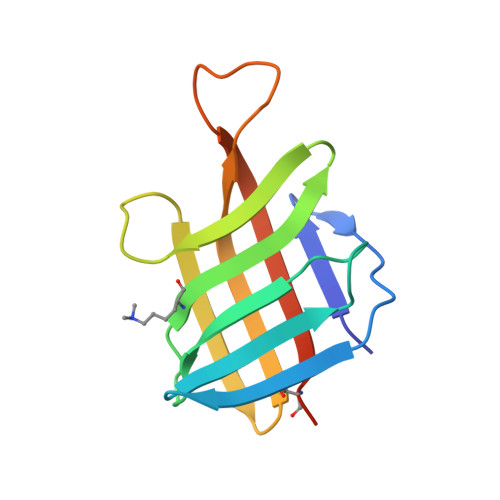 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 |  | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 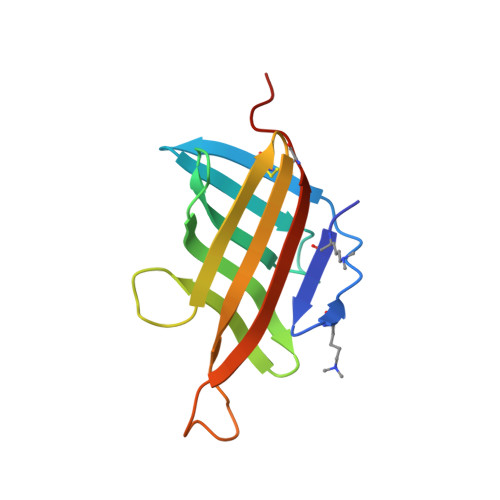 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 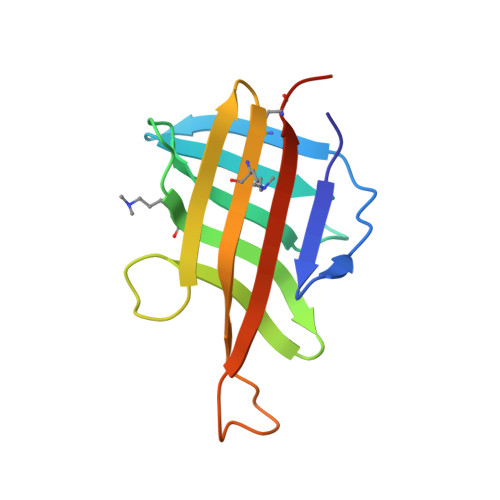 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipoprotein LpqH | 119 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: lpqH, Rv3763, MTV025.111 | 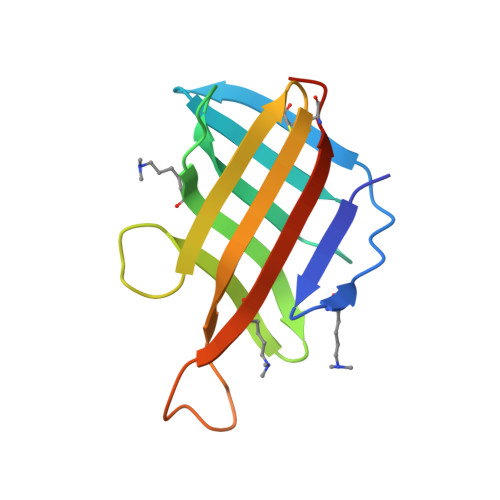 | |
UniProt | |||||
Find proteins for P9WK61 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WK61 Go to UniProtKB: P9WK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | AA [auth G] BA [auth G] H [auth A] I [auth A] L [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | J [auth A], K [auth A], Q [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | U [auth D] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MLY Query on MLY | A | L-PEPTIDE LINKING | C8 H18 N2 O2 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 100.37 | α = 90 |
| b = 100.37 | β = 90 |
| c = 239.15 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| MR-Rosetta | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 23616-002-06 F3:02 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | TBSGC R01 (A1068135) |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | TBSGC P01 (AI095208) |