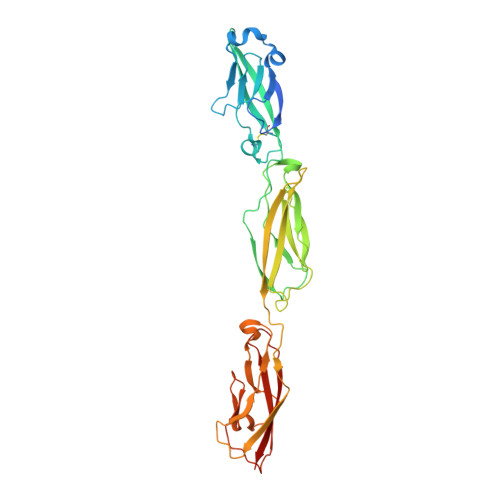
Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Nicoludis, J.M., Lau, S.Y., Scharfe, C.P., Marks, D.S., Weihofen, W.A., Gaudet, R.(2015) Structure 23: 2087-2098
- PubMed: 26481813
- DOI: https://doi.org/10.1016/j.str.2015.09.005
- Primary Citation of Related Structures:
4ZI8, 4ZI9 - PubMed Abstract:
Clustered protocadherin (Pcdh) proteins mediate dendritic self-avoidance in neurons via specific homophilic interactions in their extracellular cadherin (EC) domains. We determined crystal structures of EC1-EC3, containing the homophilic specificity-determining region, of two mouse clustered Pcdh isoforms (PcdhγA1 and PcdhγC3) to investigate the nature of the homophilic interaction. Within the crystal lattices, we observe antiparallel interfaces consistent with a role in trans cell-cell contact. Antiparallel dimerization is supported by evolutionary correlations. Two interfaces, located primarily on EC2-EC3, involve distinctive clustered Pcdh structure and sequence motifs, lack predicted glycosylation sites, and contain residues highly conserved in orthologs but not paralogs, pointing toward their biological significance as homophilic interaction interfaces. These two interfaces are similar yet distinct, reflecting a possible difference in interaction architecture between clustered Pcdh subfamilies. These structures initiate a molecular understanding of clustered Pcdh assemblies that are required to produce functional neuronal networks.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford Street, Cambridge, MA 02138, USA.