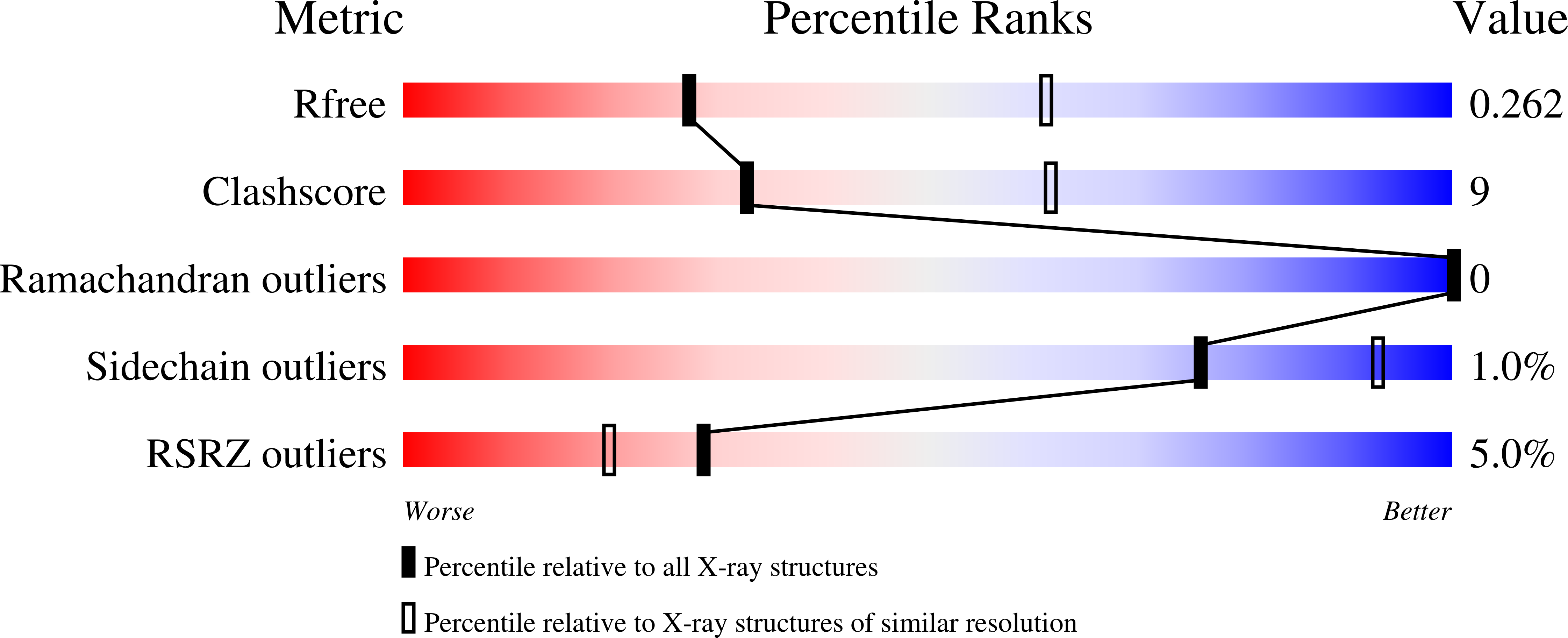
Calcium-induced conformational changes in the regulatory domain of the human mitochondrial ATP-Mg/Pi carrier.
Harborne, S.P., Ruprecht, J.J., Kunji, E.R.(2015) Biochim Biophys Acta 1847: 1245-1253
- PubMed: 26164100
- DOI: https://doi.org/10.1016/j.bbabio.2015.07.002
- Primary Citation of Related Structures:
4ZCU, 4ZCV - PubMed Abstract:
The mitochondrial ATP-Mg/Pi carrier imports adenine nucleotides from the cytosol into the mitochondrial matrix and exports phosphate. The carrier is regulated by the concentration of cytosolic calcium, altering the size of the adenine nucleotide pool in the mitochondrial matrix in response to energetic demands. The protein consists of three domains; (i) the N-terminal regulatory domain, which is formed of two pairs of fused calcium-binding EF-hands, (ii) the C-terminal mitochondrial carrier domain, which is involved in transport, and (iii) a linker region with an amphipathic α-helix of unknown function. The mechanism by which calcium binding to the regulatory domain modulates substrate transport in the carrier domain has not been resolved. Here, we present two new crystal structures of the regulatory domain of the human isoform 1. Careful analysis by SEC confirmed that although the regulatory domain crystallised as dimers, full-length ATP-Mg/Pi carrier is monomeric. Therefore, the ATP-Mg/Pi carrier must have a different mechanism of calcium regulation than the architecturally related aspartate/glutamate carrier, which is dimeric. The structure showed that an amphipathic α-helix is bound to the regulatory domain in a hydrophobic cleft of EF-hand 3/4. Detailed bioinformatics analyses of different EF-hand states indicate that upon release of calcium, EF-hands close, meaning that the regulatory domain would release the amphipathic α-helix. We propose a mechanism for ATP-Mg/Pi carriers in which the amphipathic α-helix becomes mobile upon release of calcium and could block the transport of substrates across the mitochondrial inner membrane.
Organizational Affiliation:
The Medical Research Council, Mitochondrial Biology Unit, Cambridge Biomedical Campus, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 0XY, UK.