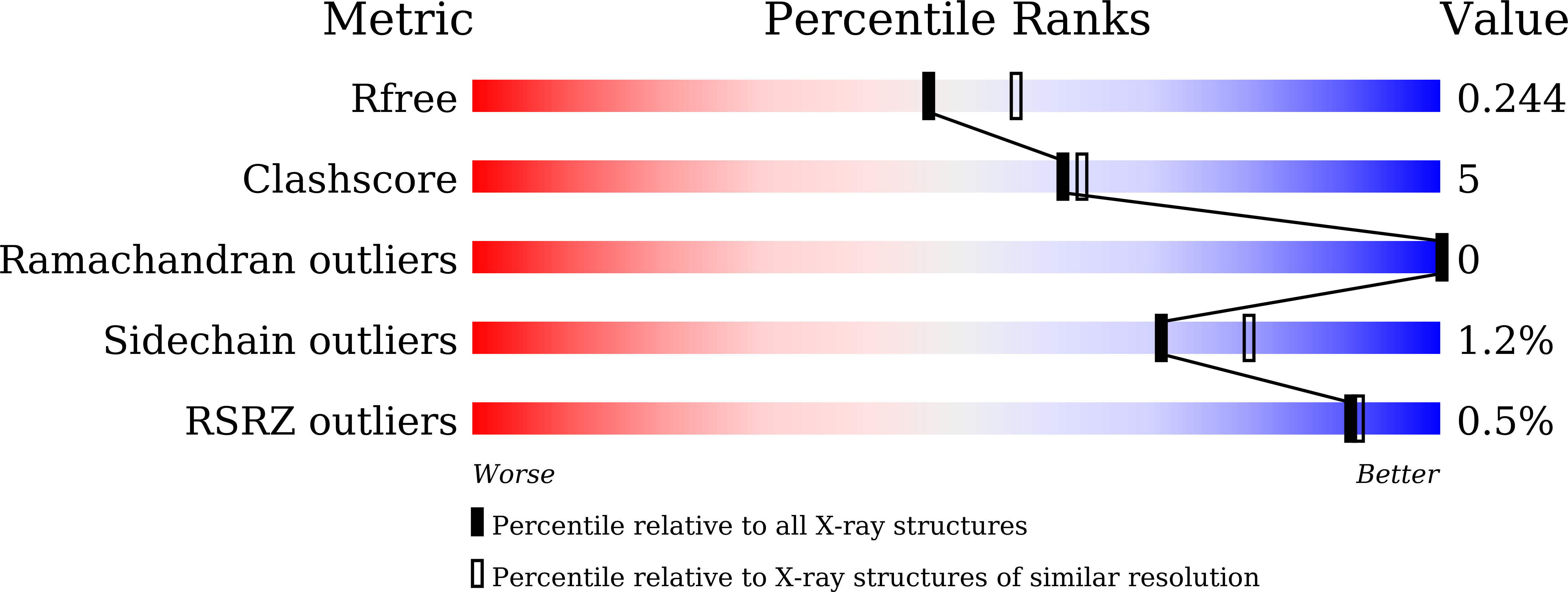
Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Zhang, F., Song, Y., Niu, L., Teng, M., Li, X.(2015) Acta Biochim Biophys Sin (Shanghai) 47: 1005-1010
- PubMed: 26508479
- DOI: https://doi.org/10.1093/abbs/gmv114
- Primary Citation of Related Structures:
4YLY - PubMed Abstract:
Peptidyl-tRNA hydrolase (Pth) catalyzes the release of tRNA to relieve peptidyl-tRNA accumulation. Because Pth activity is essential for the viability of bacteria, Pth is regarded as a promising target for the discovery of new antimicrobial agents. Here, the structure of Pth from the Gram-positive bacterium Staphylococcus aureus (SaPth) was solved by X-ray crystallography at a 2.25 Å resolution. The SaPth structure exhibits significant structural similarity with other members of the Pth superfamily, with a conserved α/β/α sandwich fold. A molecular phylogenetic analysis and a structure database search indicated that SaPth is most similar to its homolog in Streptococcus pyogenes, but it has a different substrate-binding cleft state.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, Innovation Center for Cell Signaling Network, School of Life Science, University of Science and Technology of China, Hefei 230026, China Key Laboratory of Structural Biology, Hefei Science Center, Chinese Academy of Science, Hefei 230026, China.