Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents.
Kincaid, V.A., London, N., Wangkanont, K., Wesener, D.A., Marcus, S.A., Heroux, A., Nedyalkova, L., Talaat, A.M., Forest, K.T., Shoichet, B.K., Kiessling, L.L.(2015) ACS Chem Biol 10: 2209-2218
- PubMed: 26214585
- DOI: https://doi.org/10.1021/acschembio.5b00370
- Primary Citation of Related Structures:
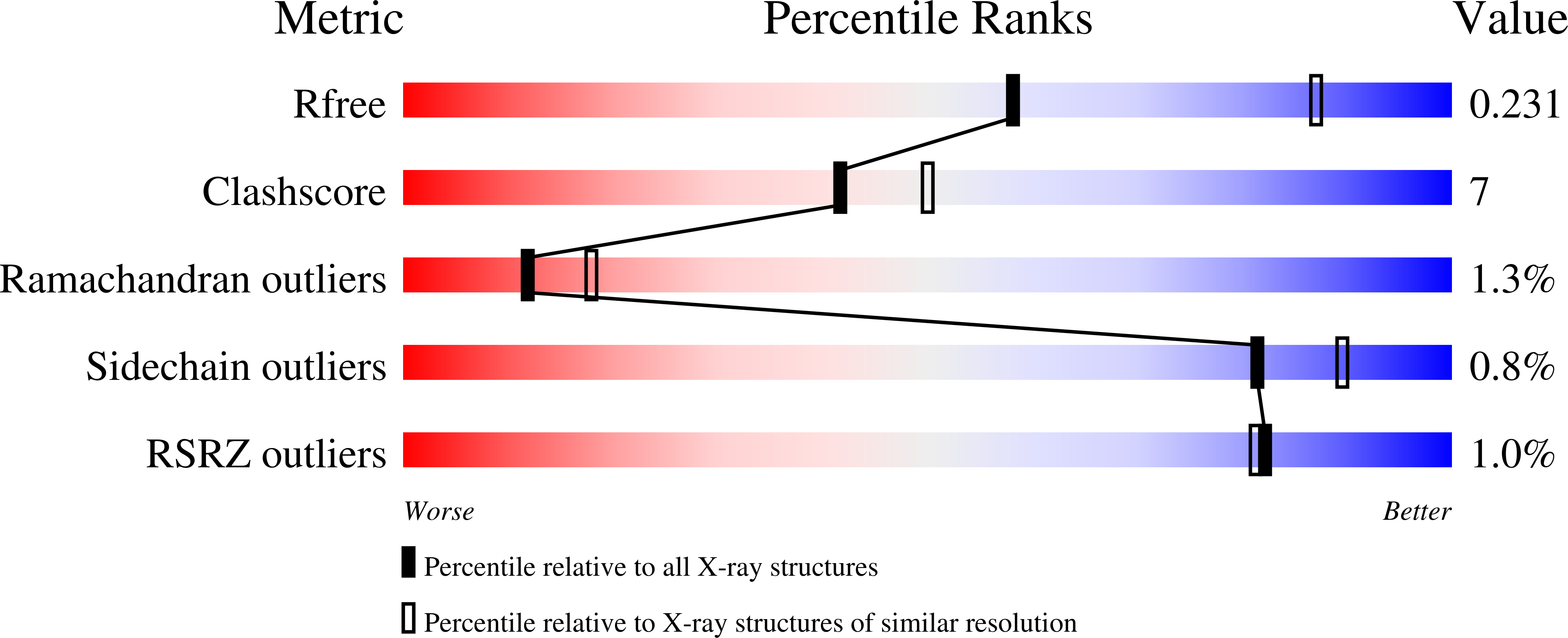
4XGK - PubMed Abstract:
Galactofuranose (Galf) is present in glycans critical for the virulence and viability of several pathogenic microbes, including Mycobacterium tuberculosis, yet the monosaccharide is absent from mammalian glycans. Uridine 5'-diphosphate-galactopyranose mutase (UGM) catalyzes the formation of UDP-Galf, which is required to produce Galf-containing glycoconjugates. Inhibitors of UGM have therefore been sought, both as antimicrobial leads and as tools to delineate the roles of Galf in cells. Obtaining cell permeable UGM probes by either design or high throughput screens has been difficult, as has elucidating how UGM binds small molecule, noncarbohydrate inhibitors. To address these issues, we employed structure-based virtual screening to uncover new inhibitor chemotypes, including a triazolothiadiazine series. These compounds are among the most potent antimycobacterial UGM inhibitors described. They also facilitated determination of a UGM-small molecule inhibitor structure, which can guide optimization. A comparison of results from the computational screen and a high-throughput fluorescence polarization (FP) screen indicated that the scaffold hits from the former had been evaluated in the FP screen but missed. By focusing on promising compounds, the virtual screen rescued false negatives, providing a blueprint for generating new UGM probes and therapeutic leads.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison , Madison, Wisconsin 53706, United States.