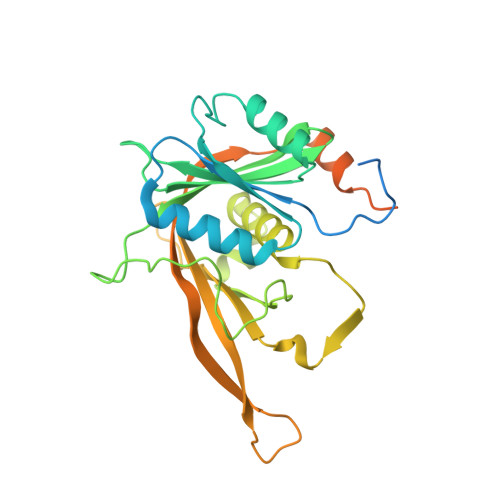
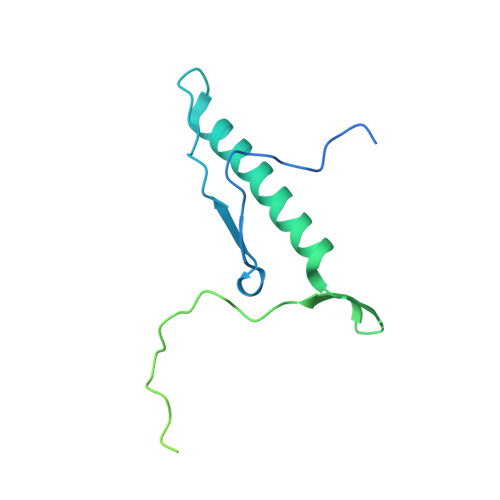
Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis.
Asano, N., Kato, K., Nakamura, A., Komoda, K., Tanaka, I., Yao, M.(2015) Nucleic Acids Res 43: 4746-4757
- PubMed: 25855814
- DOI: https://doi.org/10.1093/nar/gkv305
- Primary Citation of Related Structures:
4XD9 - PubMed Abstract:
Proteins Rpf2 and Rrs1 are required for 60S ribosomal subunit maturation. These proteins are necessary for the recruitment of three ribosomal components (5S ribosomal RNA [rRNA], RpL5 and RpL11) to the 90S ribosome precursor and subsequent 27SB pre-rRNA processing. Here we present the crystal structure of the Aspergillus nidulans (An) Rpf2-Rrs1 core complex. The core complex contains the tightly interlocked N-terminal domains of Rpf2 and Rrs1. The Rpf2 N-terminal domain includes a Brix domain characterized by similar N- and C-terminal architecture. The long α-helix of Rrs1 joins the C-terminal half of the Brix domain as if it were part of a single molecule. The conserved proline-rich linker connecting the N- and C-terminal domains of Rrs1 wrap around the side of Rpf2 and anchor the C-terminal domain of Rrs1 to a specific site on Rpf2. In addition, gel shift analysis revealed that the Rpf2-Rrs1 complex binds directly to 5S rRNA. Further analysis of Rpf2-Rrs1 mutants demonstrated that Saccharomyces cerevisiae Rpf2 R236 (corresponds to R238 of AnRpf2) plays a significant role in this binding. Based on these studies and previous reports, we have proposed a model for ribosomal component recruitment to the 90S ribosome precursor.
Organizational Affiliation:
Graduate School of Life Science, Hokkaido University, Sapporo 060-0810, Japan.