YehZYXW of Escherichia coli Is a Low-Affinity, Non-Osmoregulatory Betaine-Specific ABC Transporter.
Lang, S., Cressatti, M., Mendoza, K.E., Coumoundouros, C.N., Plater, S.M., Culham, D.E., Kimber, M.S., Wood, J.M.(2015) Biochemistry 54: 5735-5747
- PubMed: 26325238
- DOI: https://doi.org/10.1021/acs.biochem.5b00274
- Primary Citation of Related Structures:
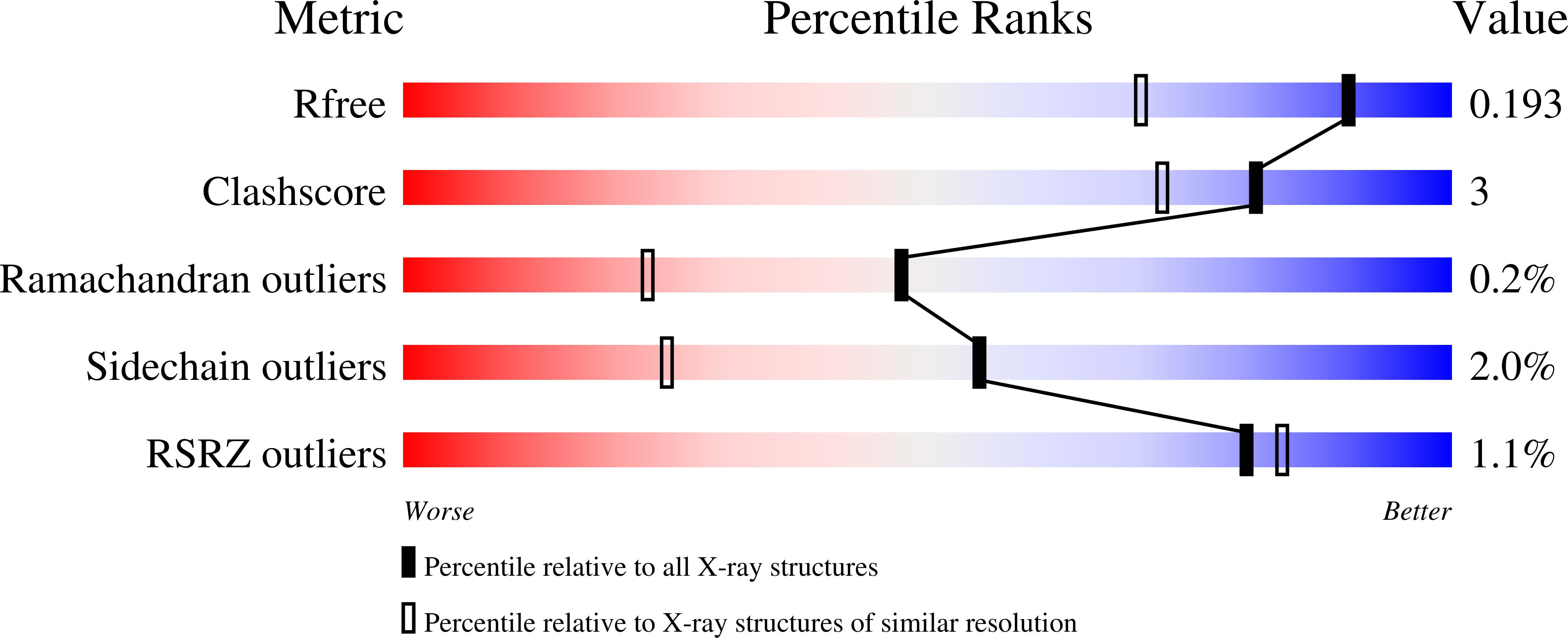
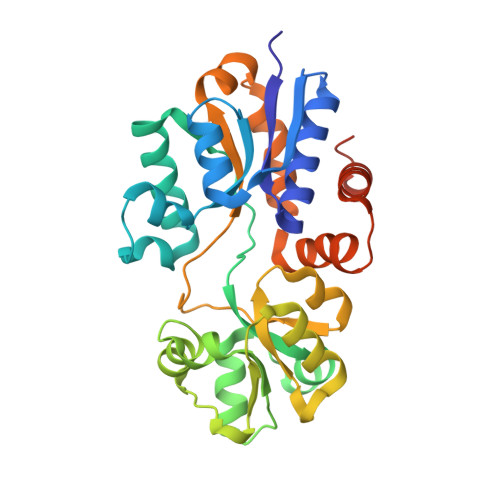
4WEP - PubMed Abstract:
Transporter-mediated osmolyte accumulation stimulates the growth of Escherichia coli in high-osmolality environments. YehZYXW was predicted to be an osmoregulatory transporter because (1) osmotic and stationary phase induction of yehZYXW is mediated by RpoS, (2) the Yeh proteins are homologous to the components of known osmoregulatory ABC transporters (e.g., ProU of E. coli), and (3) YehZ models based on the structures of periplasmic betaine-binding proteins suggested that YehZ retains key betaine-binding residues. The betaines choline-O-sulfate, glycine betaine, and dimethylsulfoniopropionate bound YehZ and ProX with millimolar and micromolar affinities, respectively, as determined by equilibrium dialysis and isothermal titration calorimetry. The crystal structure of the YehZ apoprotein, determined at 1.5 Å resolution (PDB ID: 4WEP ), confirmed its similarity to other betaine-binding proteins. Small and nonpolar residues in the hinge region of YehZ (e.g., Gly223) pack more closely than the corresponding residues in ProX, stabilizing the apoprotein. Betaines bound YehZ-Gly223Ser an order of magnitude more tightly than YehZ, suggesting that weak substrate binding in YehZ is at least partially due to apo state stabilization. Neither ProX nor YehZ bound proline. Assays based on osmoprotection or proline auxotrophy failed to detect YehZYXW-mediated uptake of proline, betaines, or other osmolytes. However, transport assays revealed low-affinity glycine betaine uptake, mediated by YehZYXW, that was inhibited at high salinity. Thus, YehZYXW is a betaine transporter that shares substrate specificity, but not an osmoregulatory function, with homologues like E. coli ProU. Other work suggests that yehZYXW may be an antivirulence locus whose expression promotes persistent, asymptomatic bacterial infection.
Organizational Affiliation:
Department of Molecular and Cellular Biology, University of Guelph , 488 Gordon Street, Guelph, ON N1G 2W1, Canada.