Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Jacewicz, A., Chico, L., Smith, P., Schwer, B., Shuman, S.(2015) RNA 21: 401-414
- PubMed: 25587180
- DOI: https://doi.org/10.1261/rna.048942.114
- Primary Citation of Related Structures:
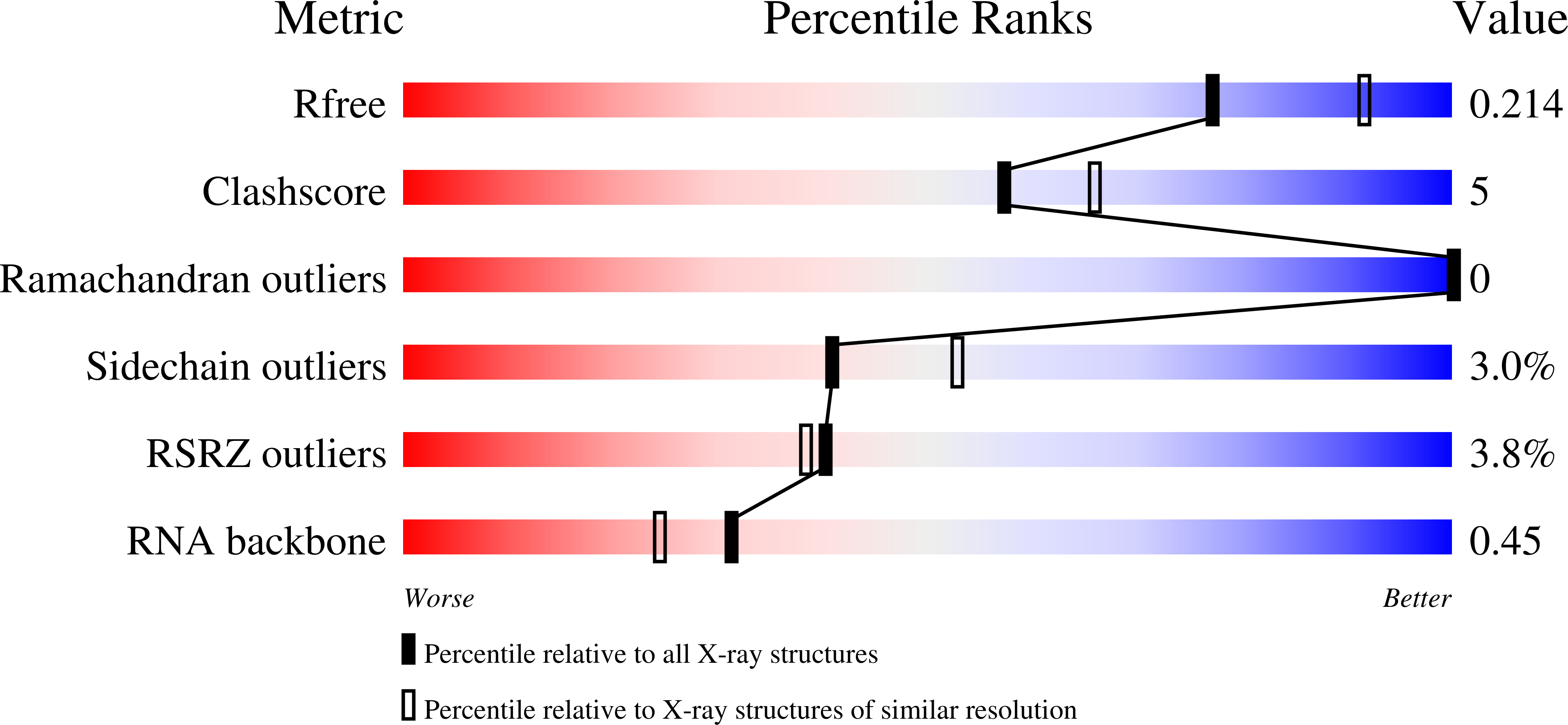
4WAL, 4WAN - PubMed Abstract:
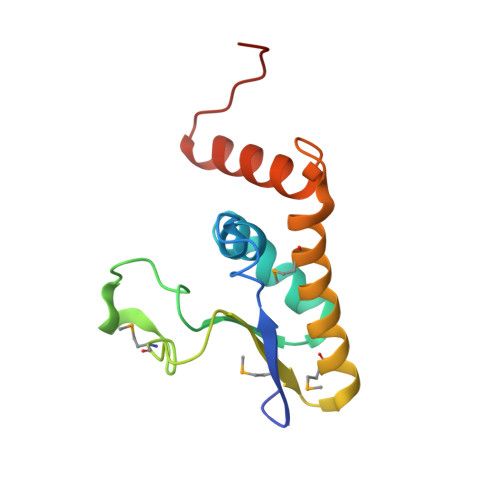
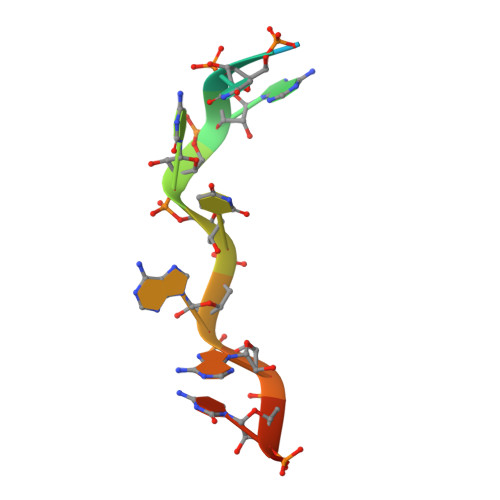
Saccharomyces cerevisiae Msl5 orchestrates spliceosome assembly by binding the intron branchpoint sequence 5'-UACUAAC and, with its heterodimer partner protein Mud2, establishing cross intron-bridging interactions with the U1 snRNP at the 5' splice site. Here we define the central Msl5 KH-QUA2 domain as sufficient for branchpoint RNA recognition. The 1.8 Å crystal structure of Msl5-(KH-QUA2) bound to the branchpoint highlights an extensive network of direct and water-mediated protein-RNA and intra-RNA atomic contacts at the interface that illuminate how Msl5 recognizes each nucleobase of the UACUAAC element. The Msl5 structure rationalizes a large body of mutational data and inspires new functional studies herein, which reveal how perturbations of the Msl5·RNA interface impede the splicing of specific yeast pre-mRNAs. We also identify interfacial mutations in Msl5 that bypass the essentiality of Sub2, a DExD-box ATPase implicated in displacing Msl5 from the branchpoint in exchange for the U2 snRNP. These studies establish an atomic resolution framework for understanding splice site selection and early spliceosome dynamics.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, New York 10065, USA.