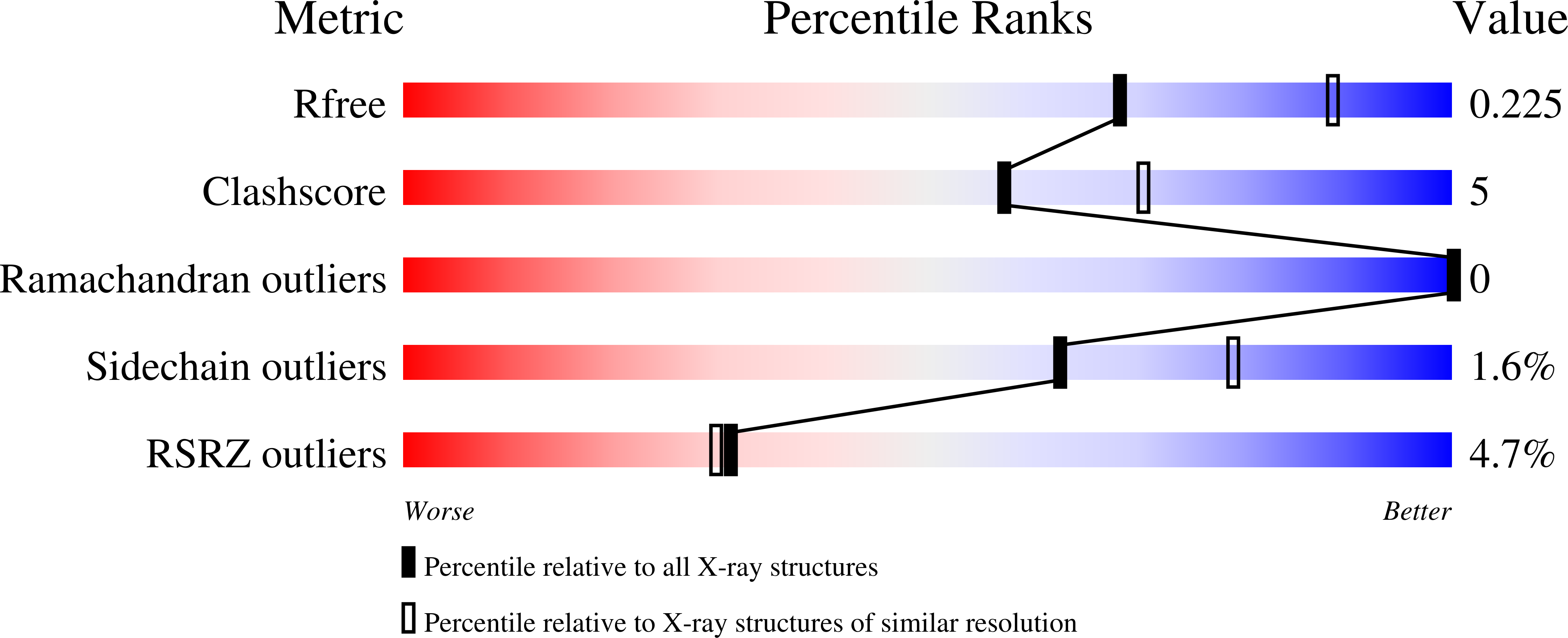
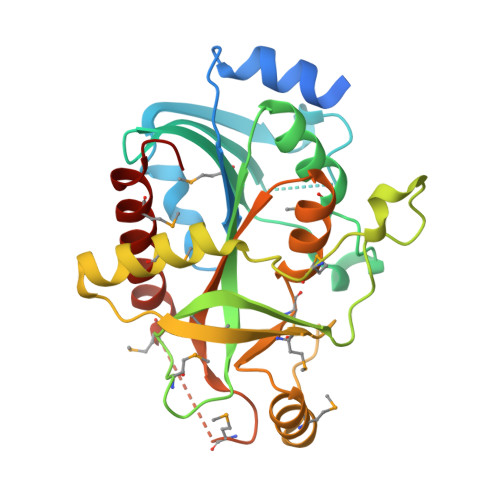
Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
Cameron, S.A., Sampathkumar, P., Ramagopal, U.A., Attonito, J., Ahmed, M., Bhosle, R., Bonanno, J., Chamala, S., Chowdhury, S., Glenn, A.S., Hammonds, J., Hillerich, B., Love, J.D., Seidel, R., Stead, M., Toro, R., Wasserman, S.R., Schramm, V.L., Almo, S.C., New York Structural Genomics Research Consortium (NYSGRC)To be published.