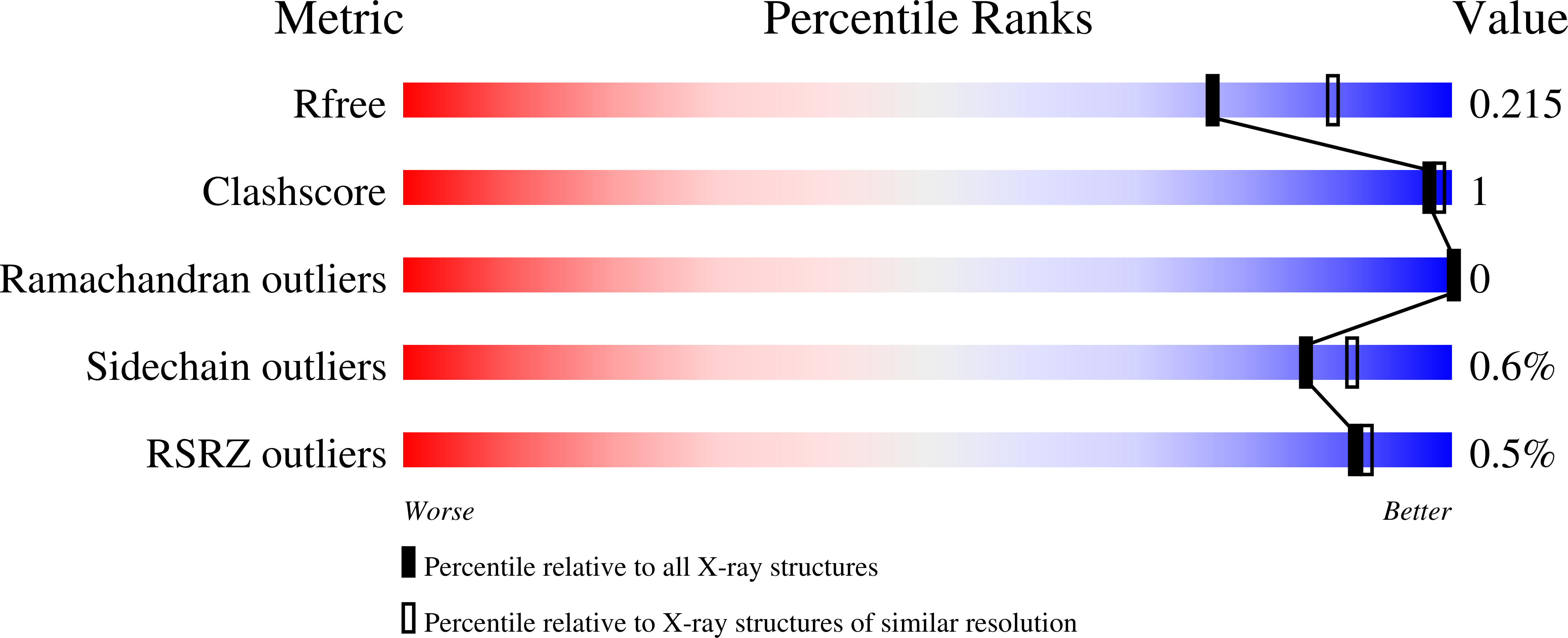
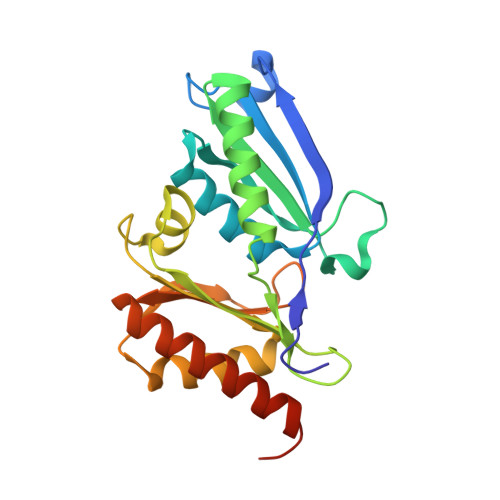
Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Thompson, M.C., Crowley, C.S., Kopstein, J., Bobik, T.A., Yeates, T.O.(2014) Acta Crystallogr F Struct Biol Commun 70: 1584-1590
- PubMed: 25484204
- DOI: https://doi.org/10.1107/S2053230X1402158X
- Primary Citation of Related Structures:
4U6I - PubMed Abstract:
The EutL shell protein is a key component of the ethanolamine-utilization microcompartment, which serves to compartmentalize ethanolamine degradation in diverse bacteria. The apparent function of this shell protein is to facilitate the selective diffusion of large cofactor molecules between the cytoplasm and the lumen of the microcompartment. While EutL is implicated in molecular-transport phenomena, the details of its function, including the identity of its transport substrate, remain unknown. Here, the 2.1 Å resolution X-ray crystal structure of a EutL shell protein bound to cobalamin (vitamin B12) is presented and the potential relevance of the observed protein-ligand interaction is briefly discussed. This work represents the first structure of a bacterial microcompartment shell protein bound to a potentially relevant cofactor molecule.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA 90095, USA.