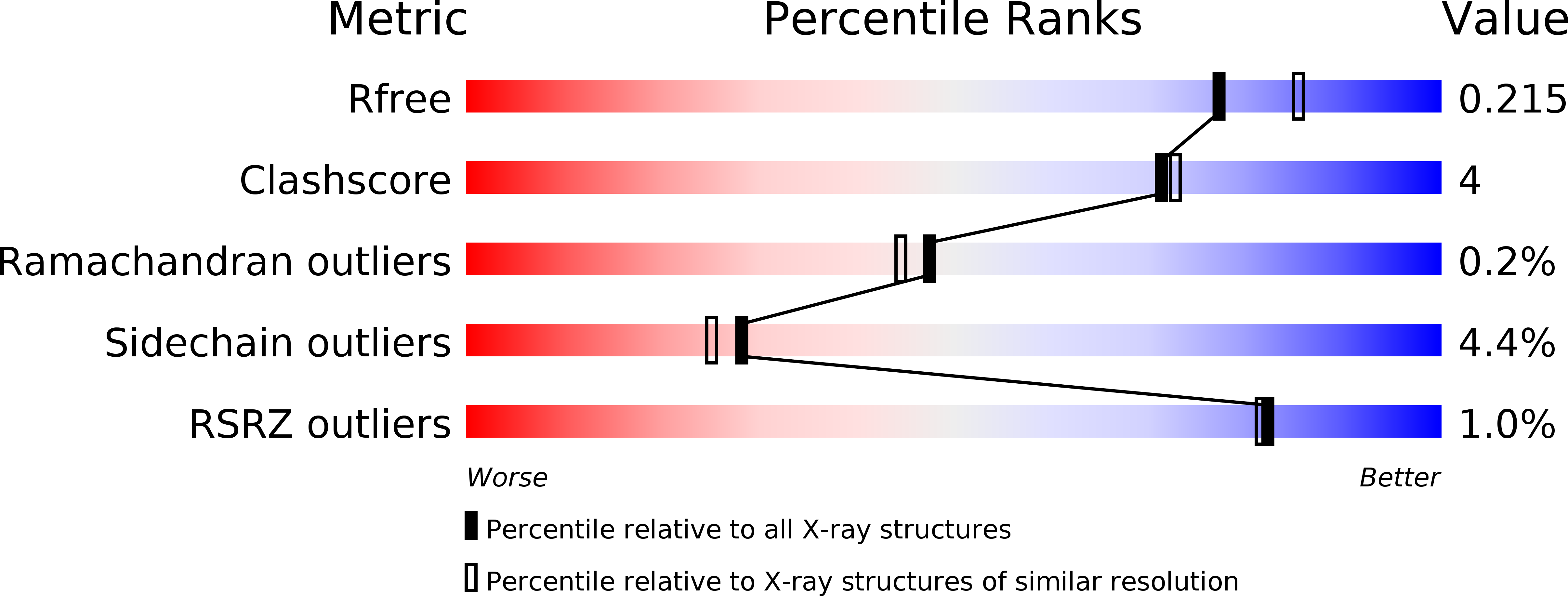
How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
Abicht, H.K., Scharer, M.A., Quade, N., Ledermann, R., Mohorko, E., Capitani, G., Hennecke, H., Glockshuber, R.(2014) J Biol Chem 289: 32431-32444
- PubMed: 25274631
- DOI: https://doi.org/10.1074/jbc.M114.607127
- Primary Citation of Related Structures:
4TXO, 4TXV - PubMed Abstract:
Two critical cysteine residues in the copper-A site (Cu(A)) on subunit II (CoxB) of bacterial cytochrome c oxidase lie on the periplasmic side of the cytoplasmic membrane. As the periplasm is an oxidizing environment as compared with the reducing cytoplasm, the prediction was that a disulfide bond formed between these cysteines must be eliminated by reduction prior to copper insertion. We show here that a periplasmic thioredoxin (TlpA) acts as a specific reductant not only for the Cu(2+) transfer chaperone ScoI but also for CoxB. The dual role of TlpA was documented best with high-resolution crystal structures of the kinetically trapped TlpA-ScoI and TlpA-CoxB mixed disulfide intermediates. They uncovered surprisingly disparate contact sites on TlpA for each of the two protein substrates. The equilibrium of CoxB reduction by TlpA revealed a thermodynamically favorable reaction, with a less negative redox potential of CoxB (E'0 = -231 mV) as compared with that of TlpA (E'0 = -256 mV). The reduction of CoxB by TlpA via disulfide exchange proved to be very fast, with a rate constant of 8.4 × 10(4) M(-1) s(-1) that is similar to that found previously for ScoI reduction. Hence, TlpA is a physiologically relevant reductase for both ScoI and CoxB. Although the requirement of ScoI for assembly of the Cu(A)-CoxB complex may be bypassed in vivo by high environmental Cu(2+) concentrations, TlpA is essential in this process because only reduced CoxB can bind copper ions.
Organizational Affiliation:
From the Institute of Molecular Biology and Biophysics and Institute of Microbiology, ETH Zürich, CH-8093 Zürich and.