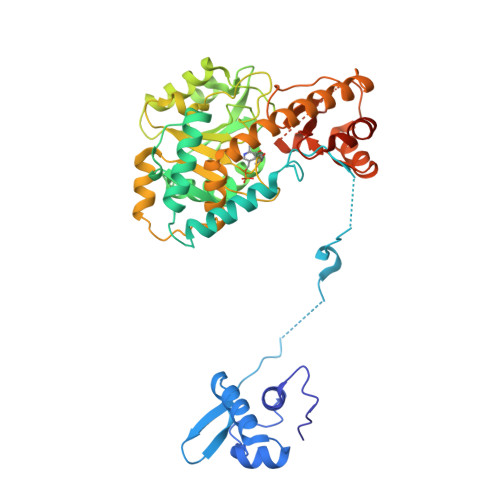
Role of the aminotransferase domain in Bacillus subtilis GabR, a pyridoxal 5'-phosphate-dependent transcriptional regulator
Okuda, K., Kato, S., Ito, T., Shiraki, S., Kawase, Y., Goto, M., Kawashima, S., Hemmi, H., Fukada, H., Yoshimura, T.(2015) Mol Microbiol 95: 245-257
- PubMed: 25388514
- DOI: https://doi.org/10.1111/mmi.12861
- Primary Citation of Related Structures:
4TV7 - PubMed Abstract:
MocR/GabR family proteins are widely distributed prokaryotic transcriptional regulators containing pyridoxal 5'-phosphate (PLP), a coenzyme form of vitamin B6. The Bacillus subtilis GabR, probably the most extensively studied MocR/GabR family protein, consists of an N-terminal DNA-binding domain and a PLP-binding C-terminal domain that has a structure homologous to aminotransferases. GabR suppresses transcription of gabR and activates transcription of gabT and gabD, which encode γ-aminobutyrate (GΑΒΑ) aminotransferase and succinate semialdehyde dehydrogenase, respectively, in the presence of PLP and GABA. In this study, we examined the mechanism underlying GabR-mediated gabTD transcription with spectroscopic, crystallographic and thermodynamic studies, focusing on the function of the aminotransferase domain. Spectroscopic studies revealed that GABA forms an external aldimine with the PLP in the aminotransferase domain. Isothermal calorimetry demonstrated that two GabR molecules bind to the 51-bp DNA fragment that contains the GabR-binding region. GABA minimally affected ΔG(binding) upon binding of GabR to the DNA fragment but greatly affected the contributions of ΔH and ΔS to ΔG(binding). GABA forms an external aldimine with PLP and causes a conformational change in the aminotransferase domain, and this change likely rearranges GabR binding to the promoter and thus activates gabTD transcription.
Organizational Affiliation:
Department of Applied Molecular Biosciences, Graduate School of Bioagricultural Sciences, Nagoya University, Frou-chou, Chikusa-ku, Nagoya, Aichi, 464-8601, Japan.