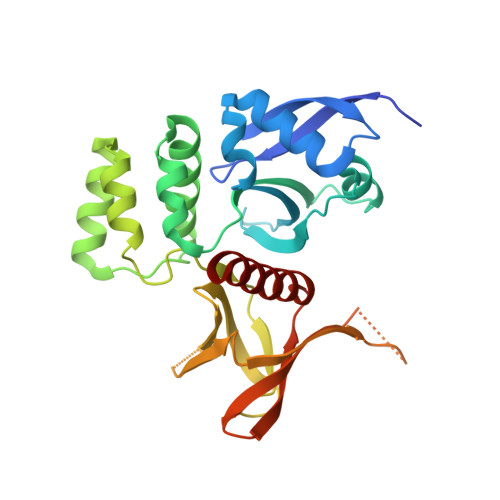
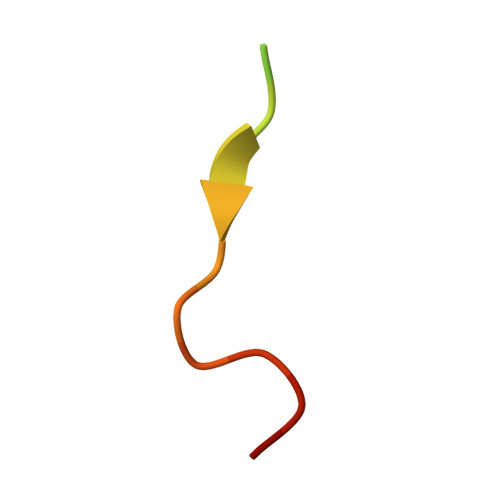
Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
Stiegler, A.L., Zhang, R., Liu, W., Boggon, T.J.(2014) J Biol Chem 289: 25362-25373
- PubMed: 25059659
- DOI: https://doi.org/10.1074/jbc.M114.584011
- Primary Citation of Related Structures:
4TKN - PubMed Abstract:
Sorting nexin 17 (SNX17) is a member of the family of cytoplasmic sorting nexin adaptor proteins that regulate endosomal trafficking of cell surface proteins. SNX17 localizes to early endosomes where it directly binds NPX(Y/F) motifs in the cytoplasmic tails of its target receptors to mediate their rates of endocytic internalization, recycling, and/or degradation. SNX17 has also been implicated in mediating cell signaling and can interact with cytoplasmic proteins. KRIT1 (Krev interaction trapped 1), a cytoplasmic adaptor protein associated with cerebral cavernous malformations, has previously been shown to interact with SNX17. Here, we demonstrate that SNX17 indeed binds directly to KRIT1 and map the binding to the second Asn-Pro-Xaa-Tyr/Phe (NPX(Y/F)) motif in KRIT1. We further characterize the interaction as being mediated by the FERM domain of SNX17. We present the co-crystal structure of SNX17-FERM with the KRIT1-NPXF2 peptide to 3.0 Å resolution and demonstrate that the interaction is highly similar in structure and binding affinity to that between SNX17 and P-selectin. We verify the molecular details of the interaction by site-directed mutagenesis and pulldown assay and thereby confirm that the major binding site for SNX17 is confined to the NPXF2 motif in KRIT1. Taken together, our results verify a direct interaction between SNX17 and KRIT1 and classify KRIT1 as a SNX17 binding partner.
Organizational Affiliation:
From the Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut 06520 amy.stiegler@yale.edu.