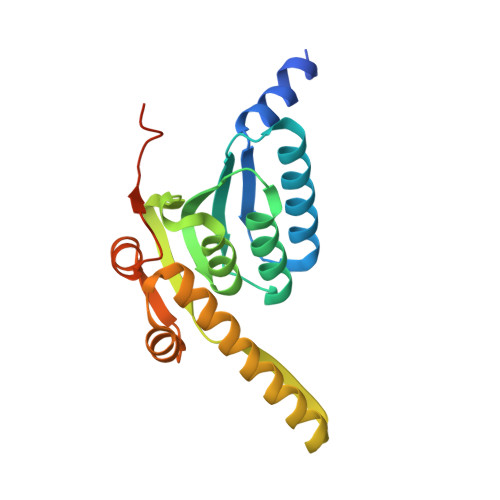
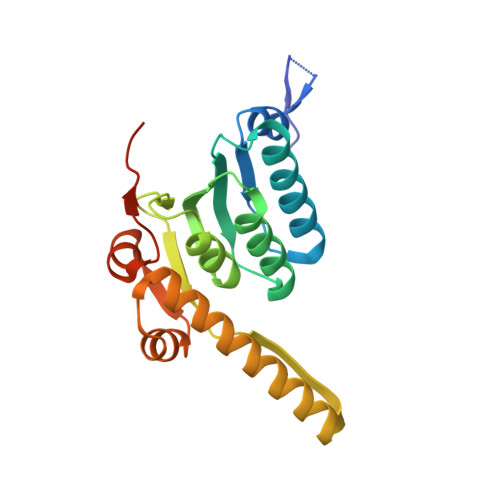
Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes.
Dahmen, M., Vielberg, M.T., Groll, M., Sieber, S.A.(2015) Angew Chem Int Ed Engl 54: 3598-3602
- PubMed: 25630955
- DOI: https://doi.org/10.1002/anie.201409325
- Primary Citation of Related Structures:
4RYF - PubMed Abstract:
Listeria monocytogenes is a devastating bacterial pathogen. Its virulence and intracellular stress tolerance are supported by caseinolytic protease P (ClpP), an enzyme that is conserved among bacteria. L. monocytogenes expresses two ClpP isoforms that are only distantly related by sequence and differ in catalysis, oligomerization, active-site composition, and N-terminal interaction sites for associated AAA(+) chaperones. The crystal structure of the ClpP1/2 heterocomplex from L. monocytogenes was solved, and in combination with biochemical studies, it provides insights into the mode of action. The results demonstrate that structural interlocking of LmClpP1 with LmClpP2 leads to the formation of a tetradecamer, aligns all 14 active sites, and enhances proteolytic activity. Furthermore, the catalytic center was identified as being responsible for the transient stability of ClpPs.
Organizational Affiliation:
Center for Integrated Protein Science Munich CIPSM, Department of Chemistry, Technische Universität München, Lichtenbergstr. 4, 85747 Garching (Germany).