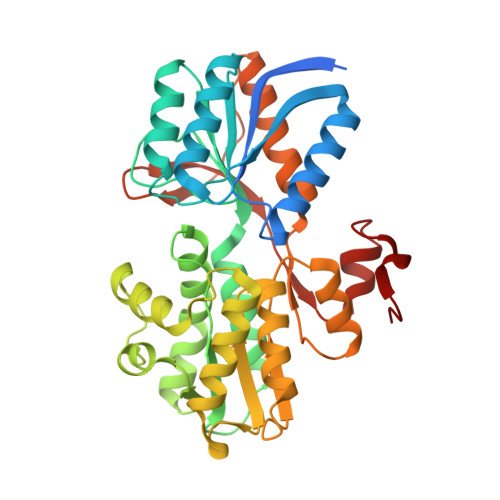
Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
Patskovsky, Y., Toro, R., Bhosle, R., Al Obaidi, N., Chamala, S., Scott Glenn, A., Attonito, J.D., Chowdhury, S., Lafleur, J., Siedel, R.D., Morisco, L.L., Wasserman, S.R., Hillerich, B., Love, J., Whalen, K.L., Gerlt, J.A., Almo, S.C.To be published.