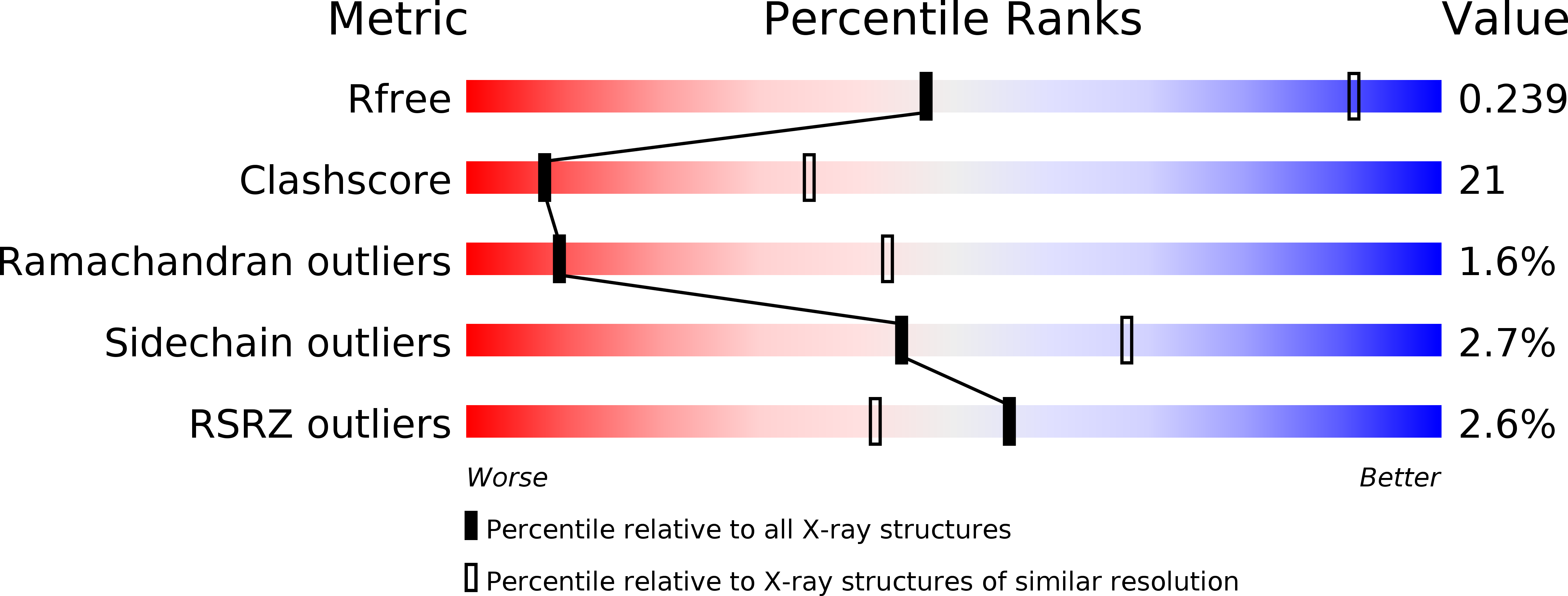
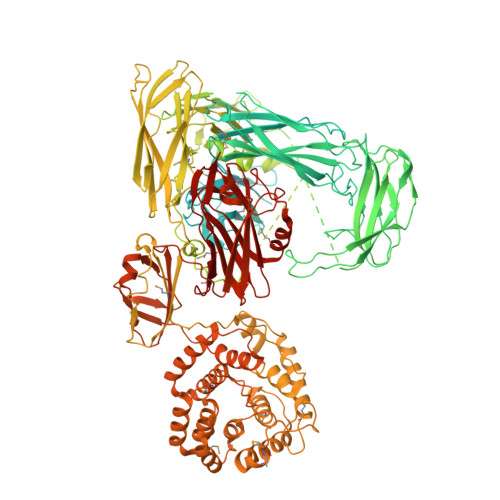
Structure of protease-cleaved Escherichia coli alpha-2-macroglobulin reveals a putative mechanism of conformational activation for protease entrapment.
Fyfe, C.D., Grinter, R., Josts, I., Mosbahi, K., Roszak, A.W., Cogdell, R.J., Wall, D.M., Burchmore, R.J., Byron, O., Walker, D.(2015) Acta Crystallogr D Biol Crystallogr 71: 1478-1486
- PubMed: 26143919
- DOI: https://doi.org/10.1107/S1399004715008548
- Primary Citation of Related Structures:
4RTD - PubMed Abstract:
Bacterial α-2-macroglobulins have been suggested to function in defence as broad-spectrum inhibitors of host proteases that breach the outer membrane. Here, the X-ray structure of protease-cleaved Escherichia coli α-2-macroglobulin is described, which reveals a putative mechanism of activation and conformational change essential for protease inhibition. In this competitive mechanism, protease cleavage of the bait-region domain results in the untethering of an intrinsically disordered region of this domain which disrupts native interdomain interactions that maintain E. coli α-2-macroglobulin in the inactivated form. The resulting global conformational change results in entrapment of the protease and activation of the thioester bond that covalently links to the attacking protease. Owing to the similarity in structure and domain architecture of Escherichia coli α-2-macroglobulin and human α-2-macroglobulin, this protease-activation mechanism is likely to operate across the diverse members of this group.
Organizational Affiliation:
Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, Scotland.