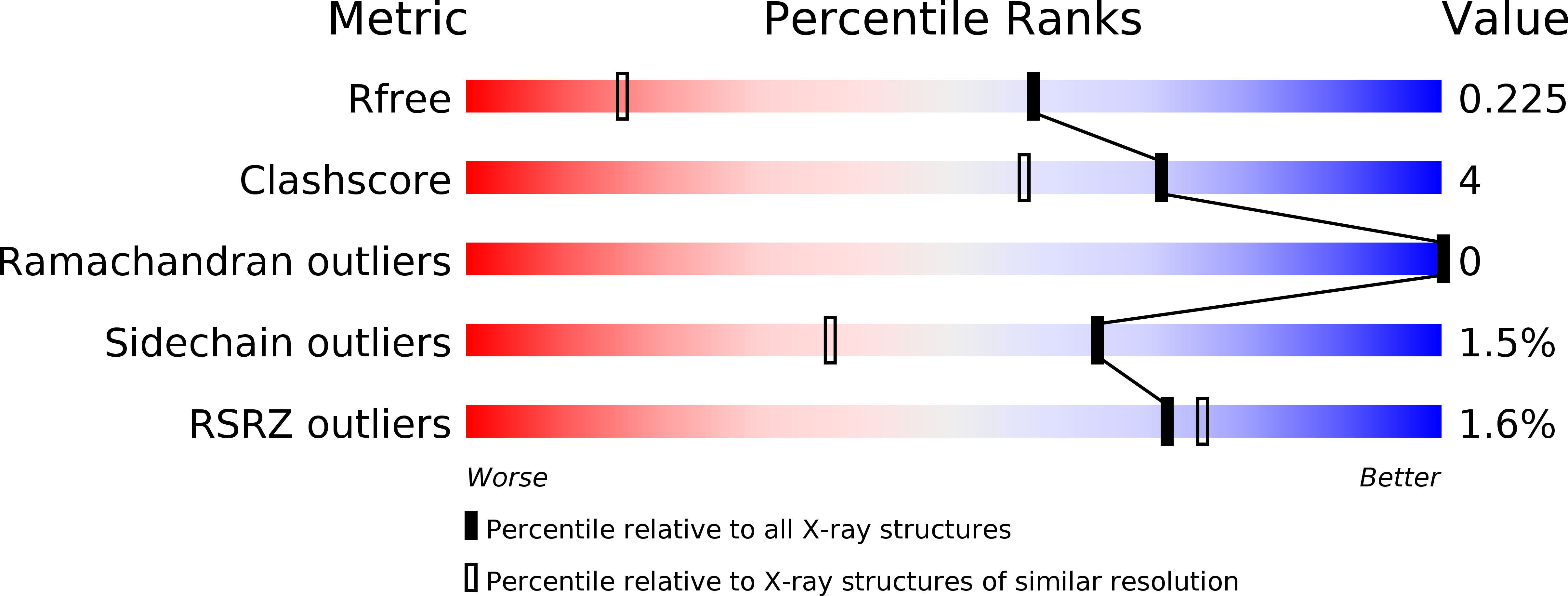
Crystal structure of dengue virus methyltransferase without S-adenosyl-L-methionine
Noble, C.G., Li, S.H., Dong, H., Chew, S.H., Shi, P.Y.(2014) Antiviral Res 111C: 78-81
- PubMed: 25241250
- DOI: https://doi.org/10.1016/j.antiviral.2014.09.003
- Primary Citation of Related Structures:
4R8R, 4R8S - PubMed Abstract:
Flavivirus methyltransferase is a genetically-validated antiviral target. Crystal structures of almost all available flavivirus methyltransferases contain S-adenosyl-L-methionine (SAM), the methyl donor molecule that co-purifies with the enzymes. This raises a possibility that SAM is an integral structural component required for the folding of dengue virus (DENV) methyltransferase. Here we exclude this possibility by solving the crystal structure of DENV methyltransferase without SAM. The SAM ligand was removed from the enzyme through a urea-mediated denaturation-and-renaturation protocol. The crystal structure of the SAM-depleted enzyme exhibits a vacant SAM-binding pocket, with a conformation identical to that of the SAM-enzyme co-crystal structure. Functionally, equivalent enzymatic activities (N-7 methylation, 2'-O methylation, and GMP-enzyme complex formation) were detected for the SAM-depleted and SAM-containing recombinant proteins. These results clearly indicate that the SAM molecule is not an essential component for the correct folding of DENV methyltransferase. Furthermore, the results imply a potential antiviral approach to search for inhibitors that can bind to the SAM-binding pocket and compete against SAM binding. To demonstrate this potential, we have soaked crystals of DENV methyltransferase without a bound SAM with the natural product Sinefungin and show that preformed crystals are capable of binding ligands in this pocket.
Organizational Affiliation:
Novartis Institute for Tropical Diseases, 10 Biopolis Road, 05-01 Chromos, Singapore 138670, Singapore.