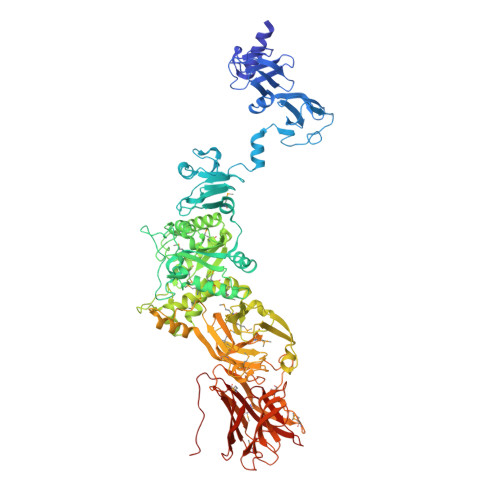
Function of bacteriophage G7C esterase tailspike in host cell adsorption.
Prokhorov, N.S., Riccio, C., Zdorovenko, E.L., Shneider, M.M., Browning, C., Knirel, Y.A., Leiman, P.G., Letarov, A.V.(2017) Mol Microbiol 105: 385-398
- PubMed: 28513100
- DOI: https://doi.org/10.1111/mmi.13710
- Primary Citation of Related Structures:
4QNL - PubMed Abstract:
Bacteriophages recognize and bind to their hosts with the help of receptor-binding proteins (RBPs) that emanate from the phage particle in the form of fibers or tailspikes. RBPs show a great variability in their shapes, sizes, and location on the particle. Some RBPs are known to depolymerize surface polysaccharides of the host while others show no enzymatic activity. Here we report that both RBPs of podovirus G7C - tailspikes gp63.1 and gp66 - are essential for infection of its natural host bacterium E. coli 4s that populates the equine intestinal tract. We characterize the structure and function of gp63.1 and show that unlike any previously described RPB, gp63.1 deacetylates surface polysaccharides of E. coli 4s leaving the backbone of the polysaccharide intact. We demonstrate that gp63.1 and gp66 form a stable complex, in which the N-terminal part of gp66 serves as an attachment site for gp63.1 and anchors the gp63.1-gp66 complex to the G7C tail. The esterase domain of gp63.1 as well as domains mediating the gp63.1-gp66 interaction is widespread among all three families of tailed bacteriophages.
Organizational Affiliation:
Research Center of Biotechnology, Russian Academy of Sciences, Winogradsky Institute of Microbiology, 7b2 pr. 60-letiya Oktyabrya, Moscow, 117312, Russia.