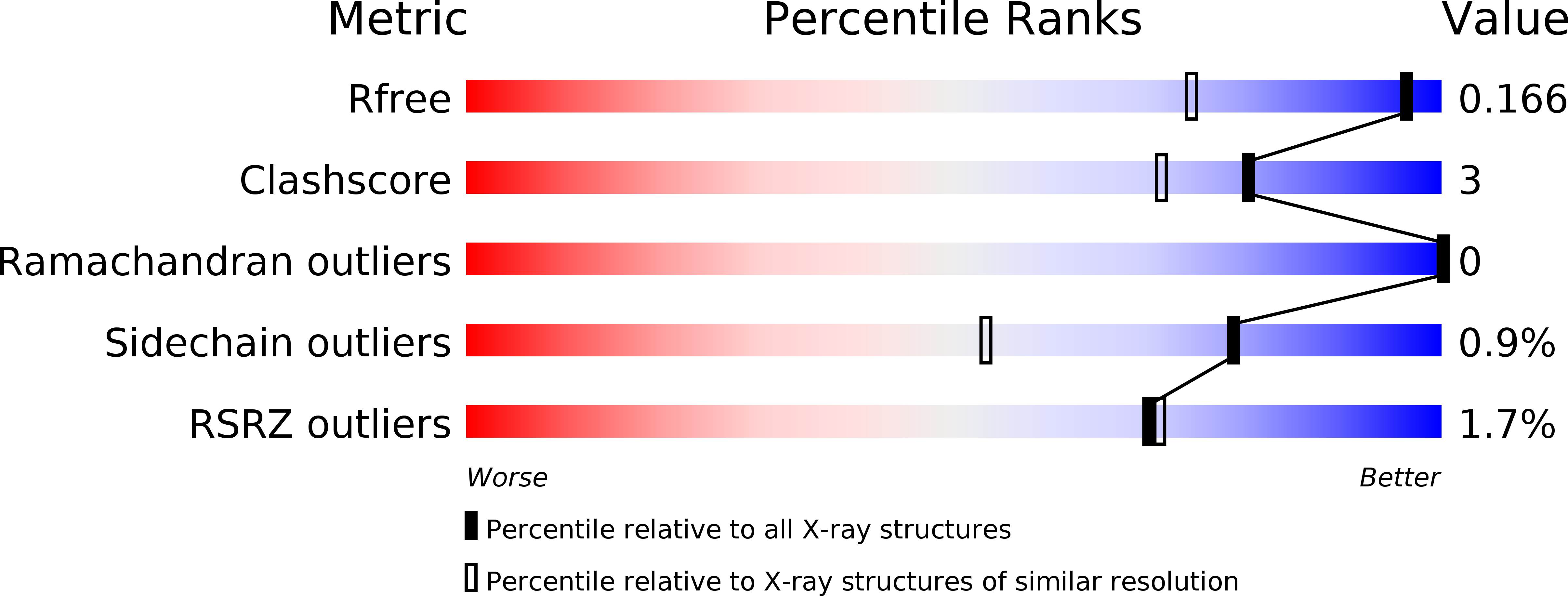
Structure of the bifunctional aminoglycoside-resistance enzyme AAC(6')-Ie-APH(2'')-Ia revealed by crystallographic and small-angle X-ray scattering analysis.
Smith, C.A., Toth, M., Weiss, T.M., Frase, H., Vakulenko, S.B.(2014) Acta Crystallogr D Biol Crystallogr 70: 2754-2764
- PubMed: 25286858
- DOI: https://doi.org/10.1107/S1399004714017635
- Primary Citation of Related Structures:
4QC6 - PubMed Abstract:
Broad-spectrum resistance to aminoglycoside antibiotics in clinically important Gram-positive staphylococcal and enterococcal pathogens is primarily conferred by the bifunctional enzyme AAC(6')-Ie-APH(2'')-Ia. This enzyme possesses an N-terminal coenzyme A-dependent acetyltransferase domain [AAC(6')-Ie] and a C-terminal GTP-dependent phosphotransferase domain [APH(2'')-Ia], and together they produce resistance to almost all known aminoglycosides in clinical use. Despite considerable effort over the last two or more decades, structural details of AAC(6')-Ie-APH(2'')-Ia have remained elusive. In a recent breakthrough, the structure of the isolated C-terminal APH(2'')-Ia enzyme was determined as the binary Mg2GDP complex. Here, the high-resolution structure of the N-terminal AAC(6')-Ie enzyme is reported as a ternary kanamycin/coenzyme A abortive complex. The structure of the full-length bifunctional enzyme has subsequently been elucidated based upon small-angle X-ray scattering data using the two crystallographic models. The AAC(6')-Ie enzyme is joined to APH(2'')-Ia by a short, predominantly rigid linker at the N-terminal end of a long α-helix. This α-helix is in turn intrinsically associated with the N-terminus of APH(2'')-Ia. This structural arrangement supports earlier observations that the presence of the intact α-helix is essential to the activity of both functionalities of the full-length AAC(6')-Ie-APH(2'')-Ia enzyme.
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource, Stanford University, Menlo Park, CA 94025, USA.