The phosphatase mechanism of bifunctional kinase/phosphatase AceK.
Wang, S., Shen, Q., Chen, G., Zheng, J., Tan, H., Jia, Z.(2014) Chem Commun (Camb) 50: 14117-14120
- PubMed: 25272278
- DOI: https://doi.org/10.1039/c4cc05375c
- Primary Citation of Related Structures:
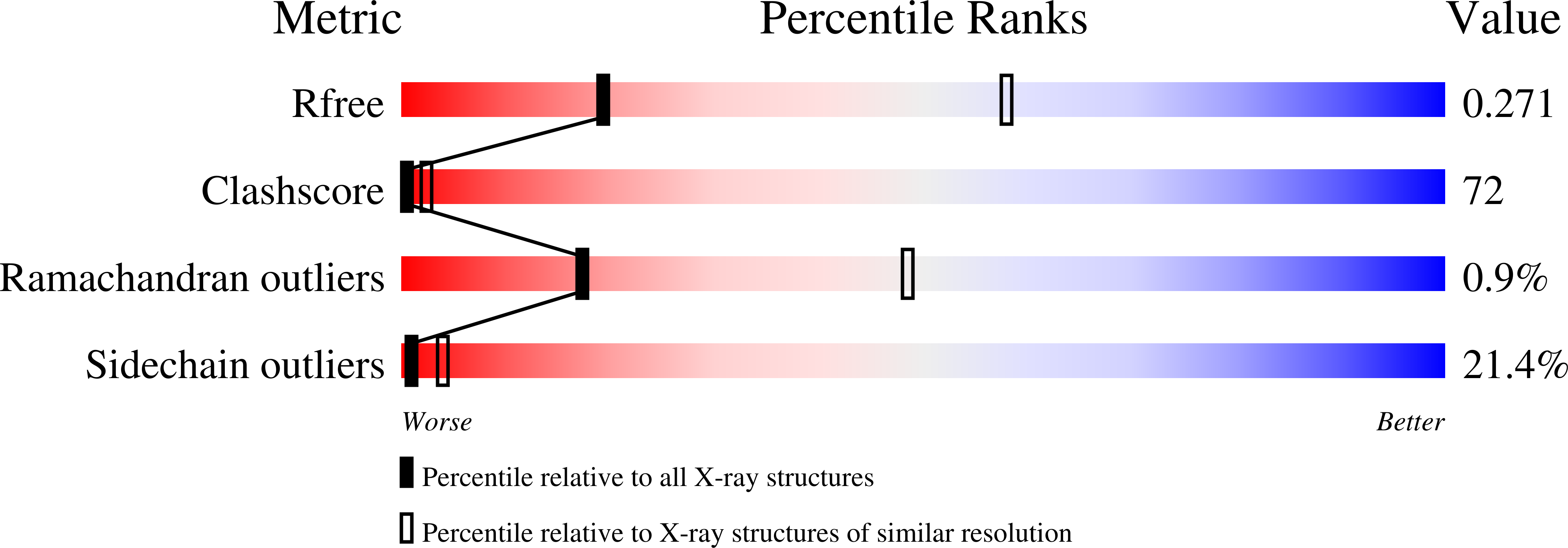
4P69 - PubMed Abstract:
We have revealed that bifunctional AceK kinase/phosphatase utilizes a stepwise addition-elimination mechanism in its dephosphorylation reaction. This work explains how AceK enables opposite kinase and phosphatase activities with Asp477 and a single Mg(2+) ion.
Organizational Affiliation:
College of Chemistry, Beijing Normal University, Beijing, 100875, China. jimin_z@bnu.edu.cn hongwei.tan@bnu.edu.cn.