Crystal structure of the bacterial A1408C-mutant ribosomal decoding site in complex with geneticin
Kondo, J., Koganei, M.To be published.
Experimental Data Snapshot
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-R(*UP*UP*GP*CP*GP*UP*CP*CP*CP*GP*(5BU)P*CP*GP*AP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3' | 23 | synthetic construct | 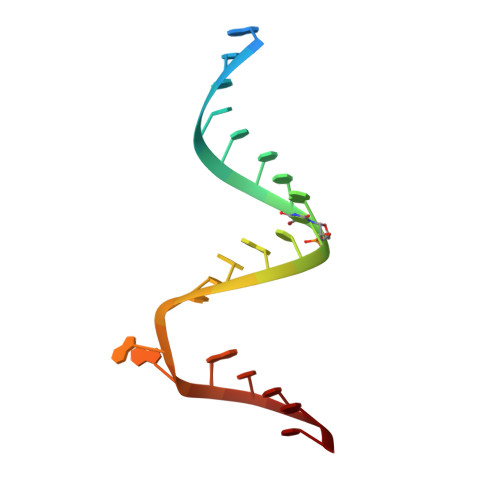 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GET Query on GET | C [auth A], D [auth A], E [auth A] | GENETICIN C20 H40 N4 O10 BRZYSWJRSDMWLG-DJWUNRQOSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 34.09 | α = 90 |
| b = 88.83 | β = 90 |
| c = 46.61 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| PHENIX | phasing |
| CNS | refinement |
| PDB_EXTRACT | data extraction |
| SOLVE | phasing |
| d*TREK | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| MEXT | Japan | 23790054 |