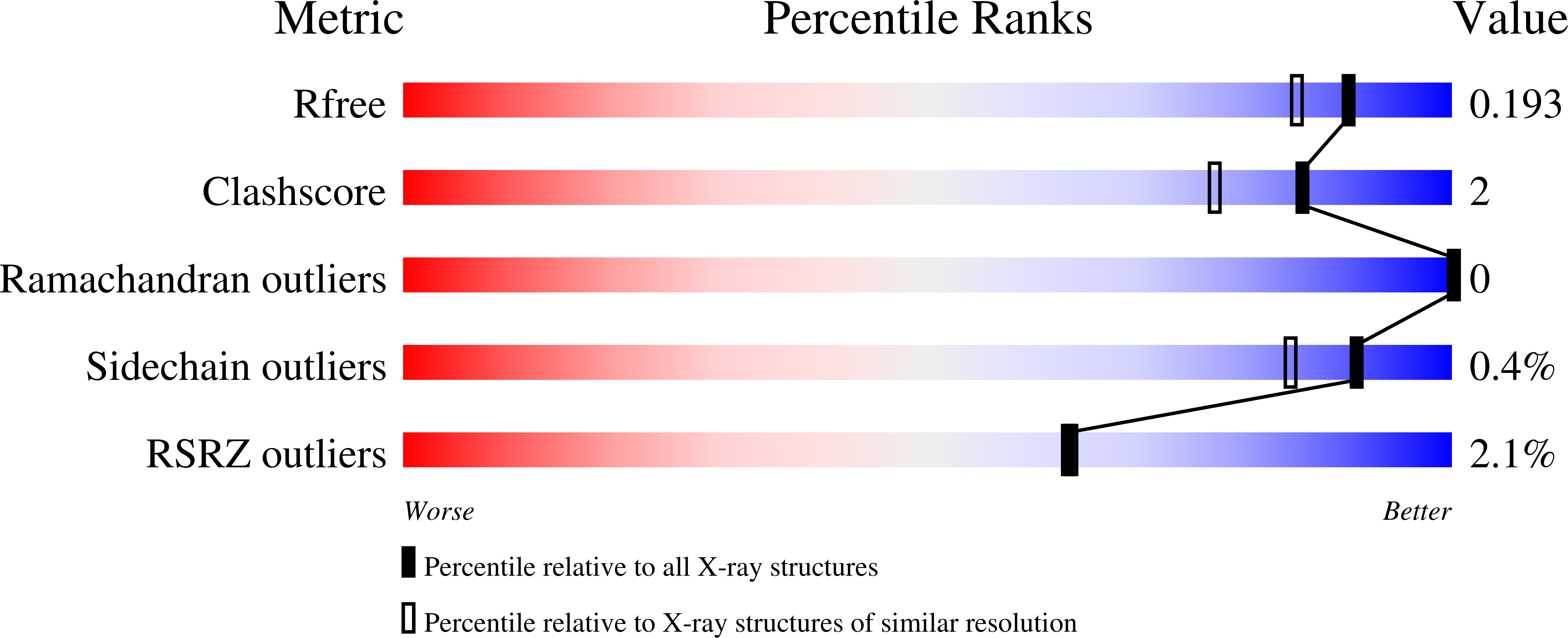
The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
Gheorghita, A.A., Wolfram, F., Whitfield, G.B., Jacobs, H.M., Pfoh, R., Wong, S.S.Y., Guitor, A.K., Goodyear, M.C., Berezuk, A.M., Khursigara, C.M., Parsek, M.R., Howell, P.L.(2022) J Biol Chem 298: 101560-101560
- PubMed: 34990713
- DOI: https://doi.org/10.1016/j.jbc.2021.101560
- Primary Citation of Related Structures:
4OZV, 4OZW, 7SA8 - PubMed Abstract:
Pseudomonas aeruginosa is an opportunistic human pathogen and a leading cause of chronic infection in the lungs of individuals with cystic fibrosis. After colonization, P. aeruginosa often undergoes a phenotypic conversion to mucoidy, characterized by overproduction of the alginate exopolysaccharide. This conversion is correlated with poorer patient prognoses. The majority of genes required for alginate synthesis, including the alginate lyase, algL, are located in a single operon. Previous investigations of AlgL have resulted in several divergent hypotheses regarding the protein's role in alginate production. To address these discrepancies, we determined the structure of AlgL and, using multiple sequence alignments, identified key active site residues involved in alginate binding and catalysis. In vitro enzymatic analysis of active site mutants highlights R249 and Y256 as key residues required for alginate lyase activity. In a genetically engineered P. aeruginosa strain where alginate biosynthesis is under arabinose control, we found that AlgL is required for cell viability and maintaining membrane integrity during alginate production. We demonstrate that AlgL functions as a homeostasis enzyme to clear the periplasmic space of accumulated polymer. Constitutive expression of the AlgU/T sigma factor mitigates the effects of an algL deletion during alginate production, suggesting that an AlgU/T-regulated protein or proteins can compensate for an algL deletion. Together, our study demonstrates the role of AlgL in alginate biosynthesis, explains the discrepancies observed previously across other P. aeruginosa ΔalgL genetic backgrounds, and clarifies the existing divergent data regarding the function of AlgL as an alginate degrading enzyme.
Organizational Affiliation:
Program in Molecular Medicine, The Hospital for Sick Children, Toronto, Ontario, Canada; Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.