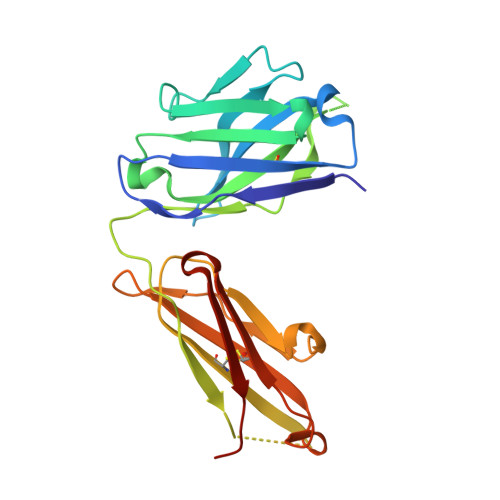
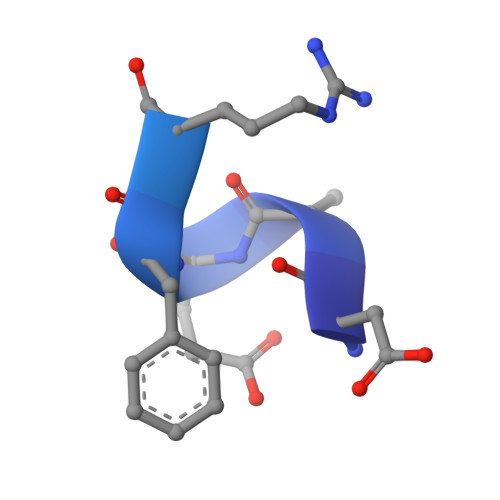
Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Feinberg, H., Saldanha, J.W., Diep, L., Goel, A., Widom, A., Veldman, G.M., Weis, W.I., Schenk, D., Basi, G.S.(2014) Alzheimers Res Ther 6: 31-31
- PubMed: 25024748
- DOI: https://doi.org/10.1186/alzrt261
- Primary Citation of Related Structures:
4ONF, 4ONG - PubMed Abstract:
Immunotherapy targeting amyloid-β peptide is under active clinical investigation for treatment of Alzheimer's disease (AD). Among the hypotheses being investigated for impact on clinical outcome are the preferred epitope or conformation of amyloid-β to target for treatment, and the mechanism of action underlying immunotherapy. Bapineuzumab (humanized 3D6), a neo-epitope specific antibody recognizing amyloid-β1-5 with strong preference for an exposed Asp residue at the N-terminus of the peptide, has undergone advanced clinical testing for treatment of AD. To gain further insight into the epitope conformation, we interrogated structural details of amino-terminal epitopes in amyloid-β using x-ray crystallography of 3D6Fab:amyloid-β complexes. Humanization of 3D6 was carried out using standard procedures integrating recombinant methods, sequence informatics, and homology modeling predictions to identify important mouse framework residues for retention in the finished humanized product. Here we report the crystal structure of a recombinant Fab fragment of 3D6 in complex with amyloid-β1-7 solved at 2.0 Å resolution. The N-terminus of amyloid-β is bound to 3D6 as a 310 helix. The amino-terminal Asp residue is buried deepest in the antibody binding pocket, with the Cβ atom of residue 6 visible at the entrance to the binding pocket near the surface of the antibody. We further evaluate homology model based predictions used to guide humanization of 3D6 to bapineuzumab, with actual structure of the Fab. The structure of the Fab:amyloid-β complex validates design of the humanized antibody, and confirms the amyloid-β epitope recognized by 3D6 as previously mapped by ELISA. The conformation of amyloid-β antigen recognized by 3D6 is novel and distinct from other antibodies recognizing N-terminal epitopes. Our result provides the first report demonstrating structural conservation of antigen contact residues, and conformation of antigen recognized, between the parent murine antibody and its humanized version.
Organizational Affiliation:
Departments of Structural Biology and of Molecular & Cellular Physiology, 299 Campus Drive, Stanford University School of Medicine, Stanford, CA 94305, USA.