Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Patra, D., Mishra, P., Surolia, A., Vijayan, M.(2014) Glycobiology 24: 956-965
- PubMed: 24957055
- DOI: https://doi.org/10.1093/glycob/cwu059
- Primary Citation of Related Structures:
4OIT, 4OIZ, 4OKC - PubMed Abstract:
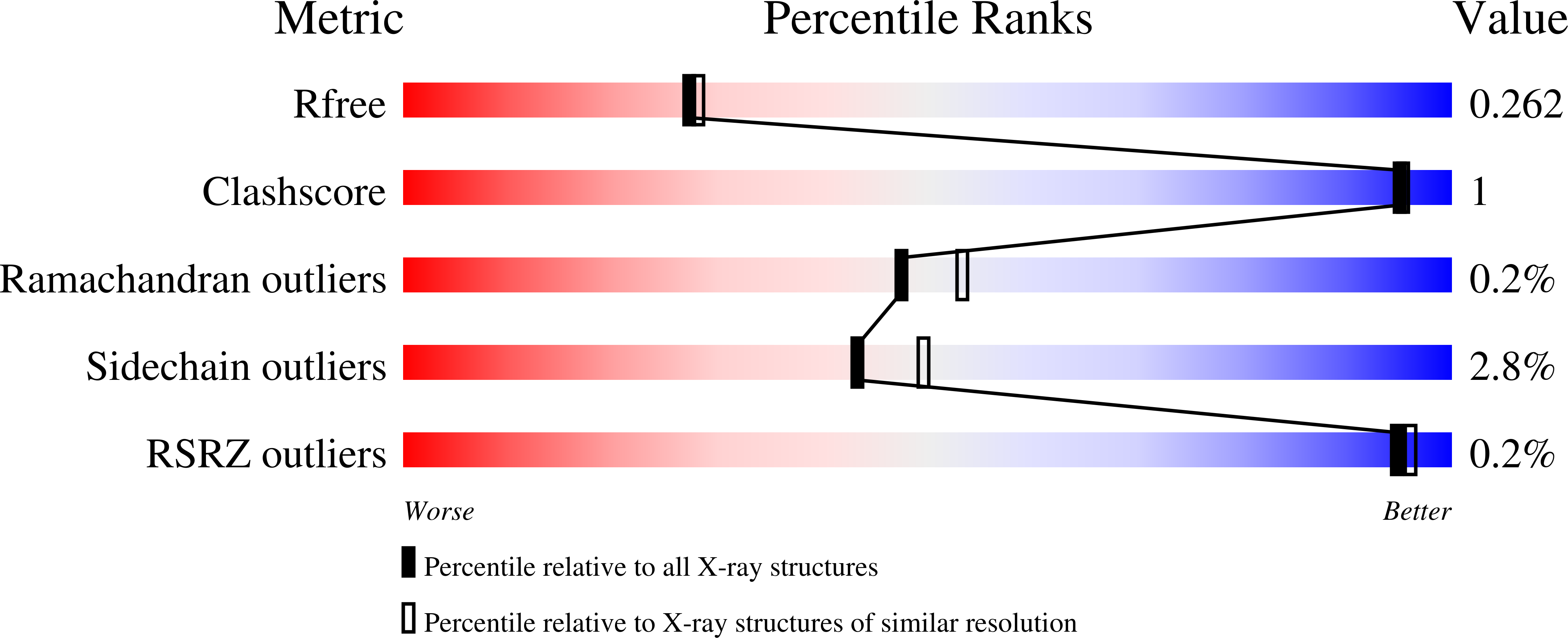
Crystal structure determination of the lectin domain of MSMEG_3662 from Mycobacterium smegmatis and its complexes with mannose and methyl-α-mannose, the first effort of its kind on a mycobacterial lectin, reveals a structure very similar to β-prism II fold lectins from plant sources, but with extensive unprecedented domain swapping in dimer formation. The two subunits in a dimer often show small differences in structure, but the two domains, not always related by 2-fold symmetry, have the same structure. Each domain carries three sugar-binding sites, similar to those in plant lectins, one on each Greek key motif. The occurrence of β-prism II fold lectins in bacteria, with characteristics similar to those from plants, indicates that this family of lectins is of ancient origin and had evolved into a mature system before bacteria and plants diverged. In plants, the number of binding sites per domain varies between one and three, whereas the number is two in the recently reported lectin domains from Pseudomonas putida and Pseudomonas aeruginosa. An analysis of the sequences of the lectins and the lectin domains shows that the level of sequence similarity among the three Greek keys in each domain has a correlation with the number of binding sites in it. Furthermore, sequence conservation among the lectins from different species is the highest for that Greek key which carries a binding site in all of them. Thus, it would appear that carbohydrate binding influences the course of the evolution of the lectin.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.