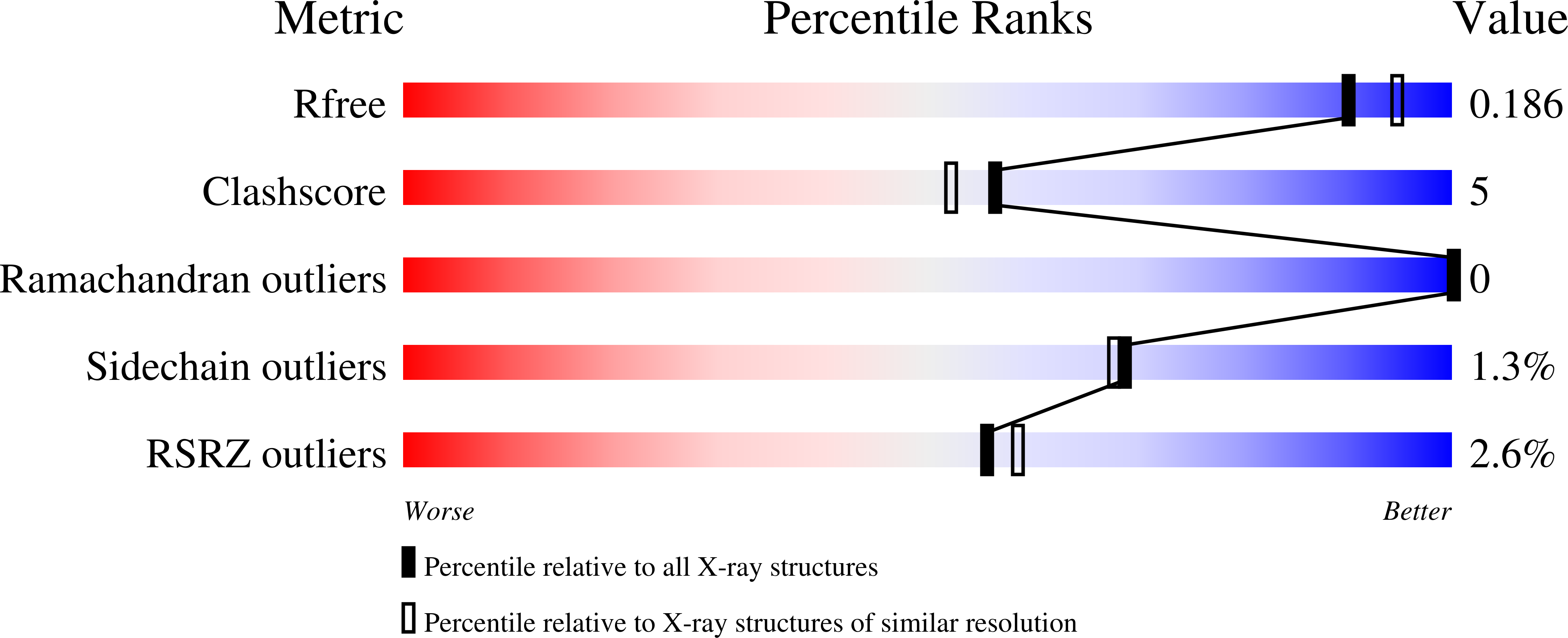
A structure-specific nucleic acid-binding domain conserved among DNA repair proteins.
Mason, A.C., Rambo, R.P., Greer, B., Pritchett, M., Tainer, J.A., Cortez, D., Eichman, B.F.(2014) Proc Natl Acad Sci U S A 111: 7618-7623
- PubMed: 24821763
- DOI: https://doi.org/10.1073/pnas.1324143111
- Primary Citation of Related Structures:
4O66 - PubMed Abstract:
SMARCAL1, a DNA remodeling protein fundamental to genome integrity during replication, is the only gene associated with the developmental disorder Schimke immuno-osseous dysplasia (SIOD). SMARCAL1-deficient cells show collapsed replication forks, S-phase cell cycle arrest, increased chromosomal breaks, hypersensitivity to genotoxic agents, and chromosomal instability. The SMARCAL1 catalytic domain (SMARCAL1(CD)) is composed of an SNF2-type double-stranded DNA motor ATPase fused to a HARP domain of unknown function. The mechanisms by which SMARCAL1 and other DNA translocases repair replication forks are poorly understood, in part because of a lack of structural information on the domains outside of the common ATPase motor. In the present work, we determined the crystal structure of the SMARCAL1 HARP domain and examined its conformation and assembly in solution by small angle X-ray scattering. We report that this domain is conserved with the DNA mismatch and damage recognition domains of MutS/MSH and NER helicase XPB, respectively, as well as with the putative DNA specificity motif of the T4 phage fork regression protein UvsW. Loss of UvsW fork regression activity by deletion of this domain was rescued by its replacement with HARP, establishing the importance of this domain in UvsW and demonstrating a functional complementarity between these structurally homologous domains. Mutation of predicted DNA-binding residues in HARP dramatically reduced fork binding and regression activities of SMARCAL1(CD). Thus, this work has uncovered a conserved substrate recognition domain in DNA repair enzymes that couples ATP-hydrolysis to remodeling of a variety of DNA structures, and provides insight into this domain's role in replication fork stability and genome integrity.
Organizational Affiliation:
Department of Biological Sciences, Vanderbilt University, Nashville, TN 37232;