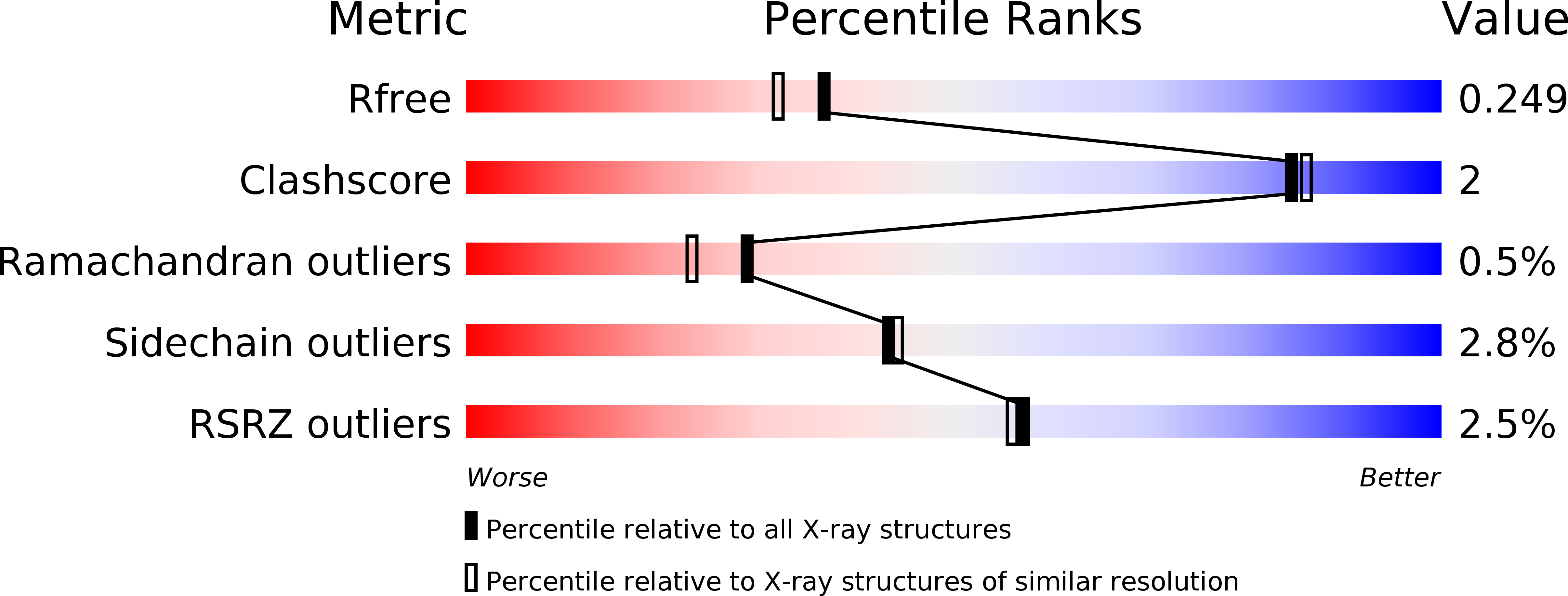
A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Peterhoff, D., Beer, B., Rajendran, C., Kumpula, E.P., Kapetaniou, E., Guldan, H., Wierenga, R.K., Sterner, R., Babinger, P.(2014) Mol Microbiol 92: 885-899
- PubMed: 24684232
- DOI: https://doi.org/10.1111/mmi.12596
- Primary Citation of Related Structures:
4JEJ, 4MM1, 4NAE, 4NAF - PubMed Abstract:
Geranylgeranylglyceryl phosphate synthase (GGGPS) family enzymes catalyse the formation of an ether bond between glycerol-1-phosphate and polyprenyl diphosphates. They are essential for the biosynthesis of archaeal membrane lipids, but also occur in bacterial species, albeit with unknown physiological function. It has been known that there exist two phylogenetic groups (I and II) of GGGPS family enzymes, but a comprehensive study has been missing. We therefore visualized the variability within the family by applying a sequence similarity network, and biochemically characterized 17 representative GGGPS family enzymes regarding their catalytic activities and substrate specificities. Moreover, we present the first crystal structures of group II archaeal and bacterial enzymes. Our analysis revealed that the previously uncharacterized bacterial enzymes from group II have GGGPS activity like the archaeal enzymes and differ from the bacterial group I enzymes that are heptaprenylglyceryl phosphate synthases. The length of the isoprenoid substrate is determined in group II GGGPS enzymes by 'limiter residues' that are different from those in group I enzymes, as shown by site-directed mutagenesis. Most of the group II enzymes form hexamers. We could disrupt these hexamers to stable and catalytically active dimers by mutating a single amino acid that acts as an 'aromatic anchor'.
Organizational Affiliation:
Institute of Biophysics and Physical Biochemistry, University of Regensburg, Regensburg, 93040, Germany.