Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Tiede, C., Tang, A.A., Deacon, S.E., Mandal, U., Nettleship, J.E., Owen, R.L., George, S.E., Harrison, D.J., Owens, R.J., Tomlinson, D.C., McPherson, M.J.(2014) Protein Eng Des Sel 27: 145-155
- PubMed: 24668773
- DOI: https://doi.org/10.1093/protein/gzu007
- Primary Citation of Related Structures:
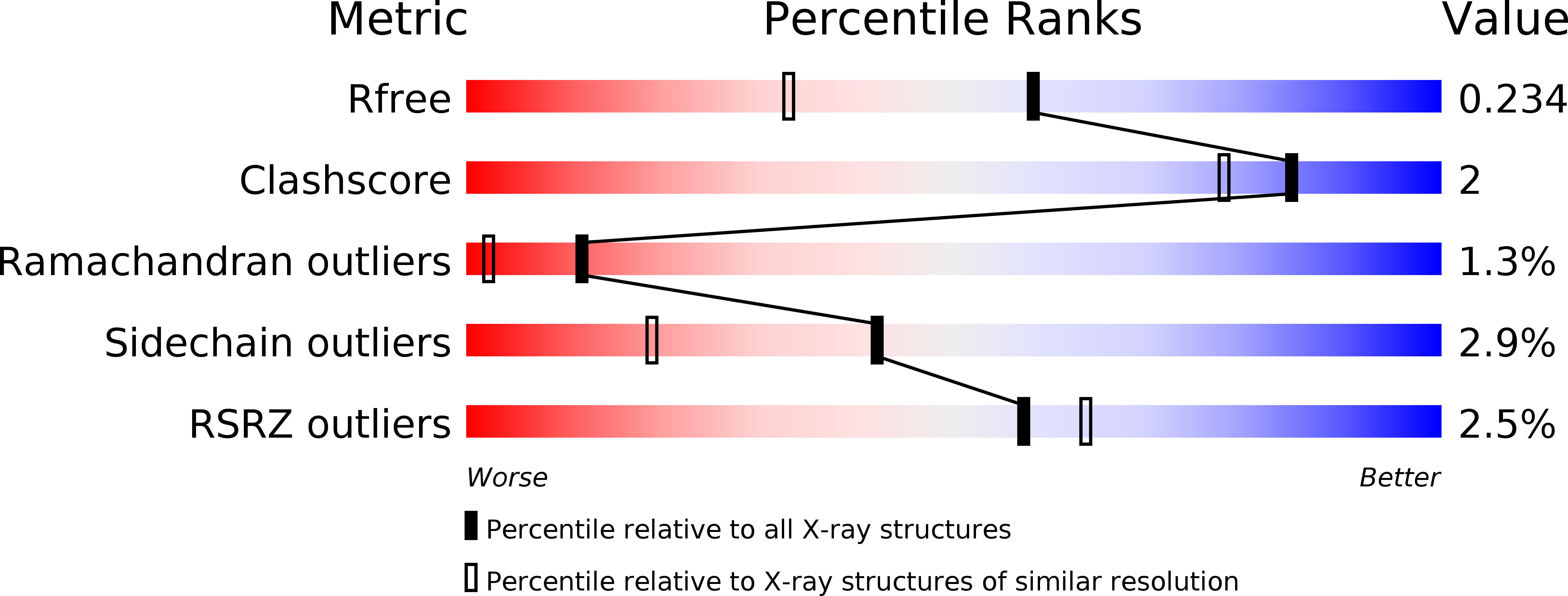
4N6T, 4N6U - PubMed Abstract:
We have designed a novel non-antibody scaffold protein, termed Adhiron, based on a phytocystatin consensus sequence. The Adhiron scaffold shows high thermal stability (Tm ca. 101°C), and is expressed well in Escherichia coli. We have determined the X-ray crystal structure of the Adhiron scaffold to 1.75 Å resolution revealing a compact cystatin-like fold. We have constructed a phage-display library in this scaffold by insertion of two variable peptide regions. The library is of high quality and complexity comprising 1.3 × 10(10) clones. To demonstrate library efficacy, we screened against the yeast Small Ubiquitin-like Modifier (SUMO). In selected clones, variable region 1 often contained sequences homologous to the known SUMO interactive motif (V/I-X-V/I-V/I). Four Adhirons were further characterised and displayed low nanomolar affinities and high specificity for yeast SUMO with essentially no cross-reactivity to human SUMO protein isoforms. We have identified binders against >100 target molecules to date including as examples, a fibroblast growth factor (FGF1), platelet endothelial cell adhesion molecule (PECAM-1; CD31), the SH2 domain Grb2 and a 12-aa peptide. Adhirons are highly stable and well expressed allowing highly specific binding reagents to be selected for use in molecular recognition applications.
Organizational Affiliation:
Biomedical Health Research Centre, BioScreening Technology Group, University of Leeds, Leeds LS2 9JT, UK.