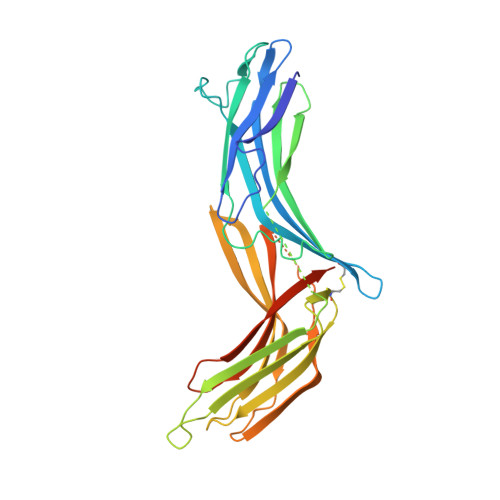
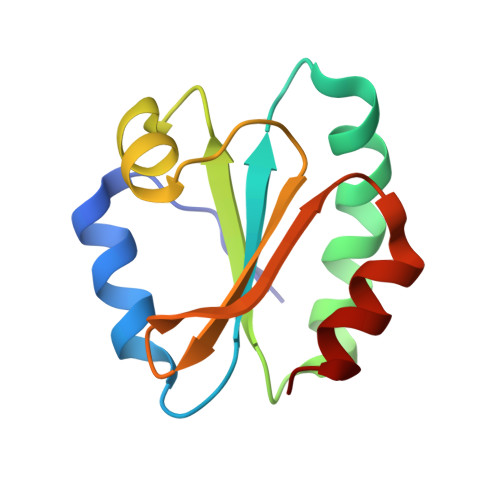
The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Hwang, J., Suh, H.W., Jeon, Y.H., Hwang, E., Nguyen, L.T., Yeom, J., Lee, S.G., Lee, C., Kim, K.J., Kang, B.S., Jeong, J.O., Oh, T.K., Choi, I., Lee, J.O., Kim, M.H.(2014) Nat Commun 5: 2958-2958
- PubMed: 24389582
- DOI: https://doi.org/10.1038/ncomms3958
- Primary Citation of Related Structures:
4GFX, 4LL1, 4LL4 - PubMed Abstract:
The redox-dependent inhibition of thioredoxin (TRX) by thioredoxin-interacting protein (TXNIP) plays a pivotal role in various cancers and metabolic syndromes. However, the molecular mechanism of this regulation is largely unknown. Here, we present the crystal structure of the TRX-TXNIP complex and demonstrate that the inhibition of TRX by TXNIP is mediated by an intermolecular disulphide interaction resulting from a novel disulphide bond-switching mechanism. Upon binding to TRX, TXNIP undergoes a structural rearrangement that involves switching of a head-to-tail interprotomer Cys63-Cys247 disulphide between TXNIP molecules to an interdomain Cys63-Cys190 disulphide, and the formation of a de novo intermolecular TXNIP Cys247-TRX Cys32 disulphide. This disulphide-switching event unexpectedly results in a domain arrangement of TXNIP that is entirely different from those of other arrestin family proteins. We further show that the intermolecular disulphide bond between TRX and TXNIP dissociates in the presence of high concentrations of reactive oxygen species. This study provides insight into TRX and TXNIP-dependent cellular regulation.
Organizational Affiliation:
1] Department of Chemistry, Korea Advanced Institute of Science and Technology, Daejeon 305-701, Korea [2] Infection and Immunity Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 305-806, Korea.