Crystal structure of ADP-ribose pyrophosphatase MutT from Rickettsia felis
Abendroth, J., Clifton, M.C., Lorimer, D., Edwards, T.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ADP-ribose pyrophosphatase MutT | 149 | Rickettsia felis URRWXCal2 | Mutation(s): 0 Gene Names: mutT, RF_0595 EC: 3.6.1 | 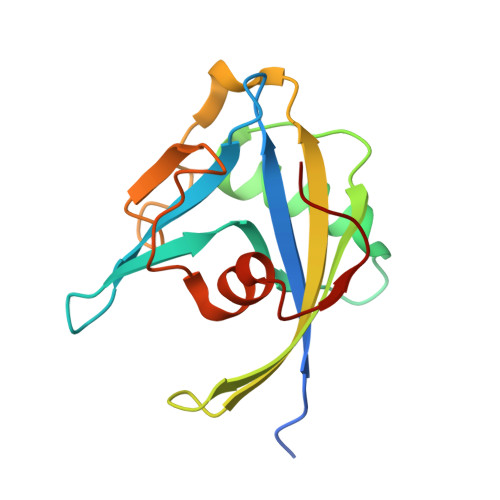 | |
UniProt | |||||
Find proteins for Q4ULX7 (Rickettsia felis (strain ATCC VR-1525 / URRWXCal2)) Explore Q4ULX7 Go to UniProtKB: Q4ULX7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q4ULX7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.98 | α = 90 |
| b = 63.78 | β = 90 |
| c = 71.72 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XSCALE | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| Blu-Ice | data collection |
| XDS | data reduction |